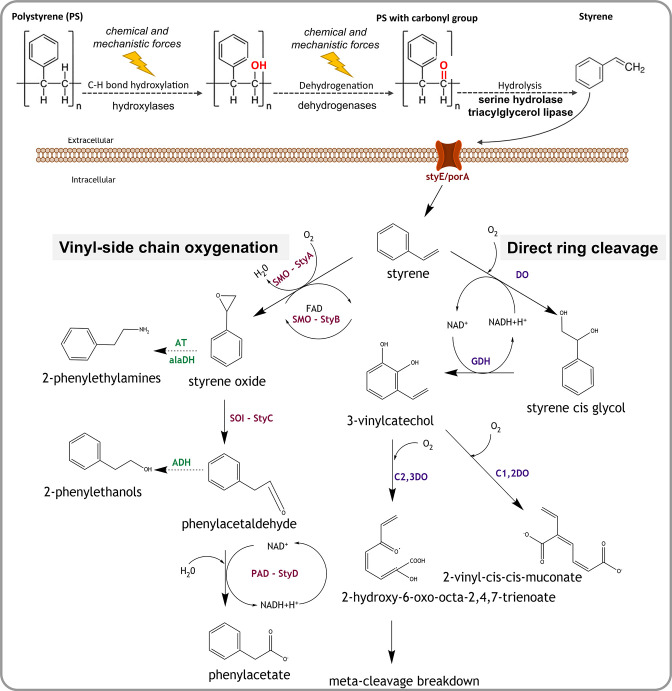
Fig. 5.
Potential pathways and enzymes of bacterial polystyrene and styrene degradation. Initially, abiotic chemical and physical factors, and potentially also extracellular enzymes such as hydroxylases and dehydrogenases, attack the polystyrene polymer creating carbonyl groups. Next, extracellular enzymes such as serine hydrolase and triacylglycerol lipase degrade the polymer to styrene monomers. A putative transmembrane protein StyE/PorA can import styrene monomers into the cell, where it is degraded via the vinyl side chain oxygenation or the direct ring cleavage pathway. In the vinyl side chain oxygenation pathway, styrene is broken down to phenylacetate using enzymes of the styABCD operon: styrene monooxygenase (StyA/SMO), styrene monooxygenase reductase component (StyB), styrene oxide isomerase (StyC/SOI) and phenylacetadehyde dehydrogenase (StyD/PAD/FeaB/ TynC). Styrene can also form 2-phenylethylamines and 2-phenylethanols as intermediates, through modifications of vinyl side chain pathway by the enzymes aminotransferase (AT), alanine dehydrogenase (alaDH) and alcohol dehydrogenase (ADH). The direct ring cleavage pathway converts styrene into Krebs cycle intermediates via meta cleavage breakdown or to muconic acid. Involved key enzymes are styrene dioxygenase (DO), 2,3-dihydrodiol dehydrogenase/cis glycol dehydrogenase (GDH), catechol-1,2-dioxygenase (C1,2DO) and catechol-2,3-dioxygenase (C2,3DO). Dashed arrows indicate hypothetical reactions.