Figure 3.
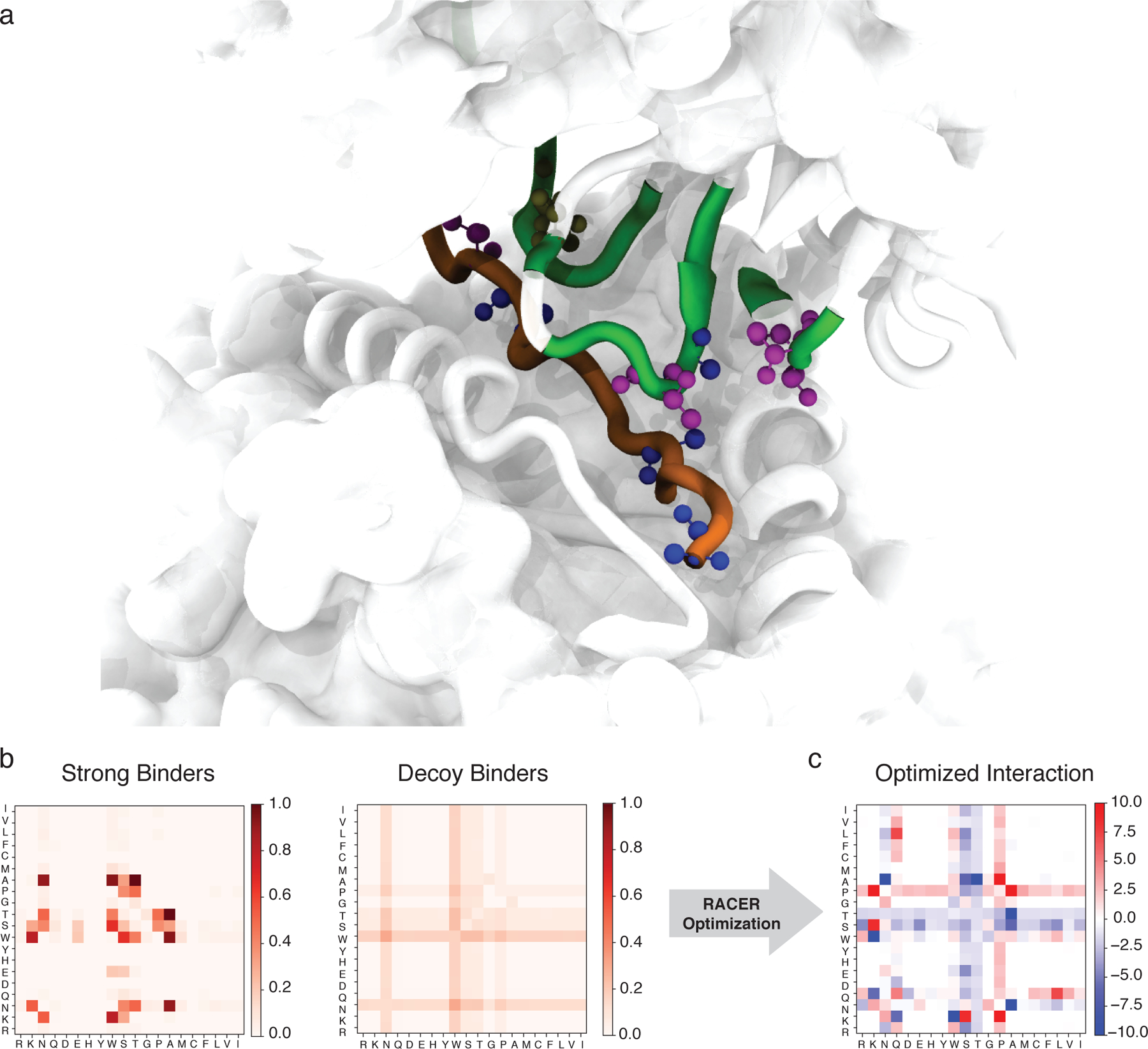
The RACER-derived energy model a. The 3D crystal structure of the 2B4 TCR bound to a specific peptide (PDBID: 3QIB). The parts of the structure that are in contact between the TCR and peptide are color-highlighted as green (TCR) and orange (peptide). Also shown are residues alanine (blue), threonine (magenta) and asparagine (tan) which are discussed in the main text (CPK representation [51]). b. The probability of contact formation (interaction set) between each two of the 20 amino acids in the set of strong binders (left) and the set of randomized decoy binders (right) of TCR 2B4. c. The residue-based interaction strength (energy model) determined by RACER for TCR 2B4. A more negative value indicates a stronger attractive interaction between the corresponding two residues.
