Figure 6.
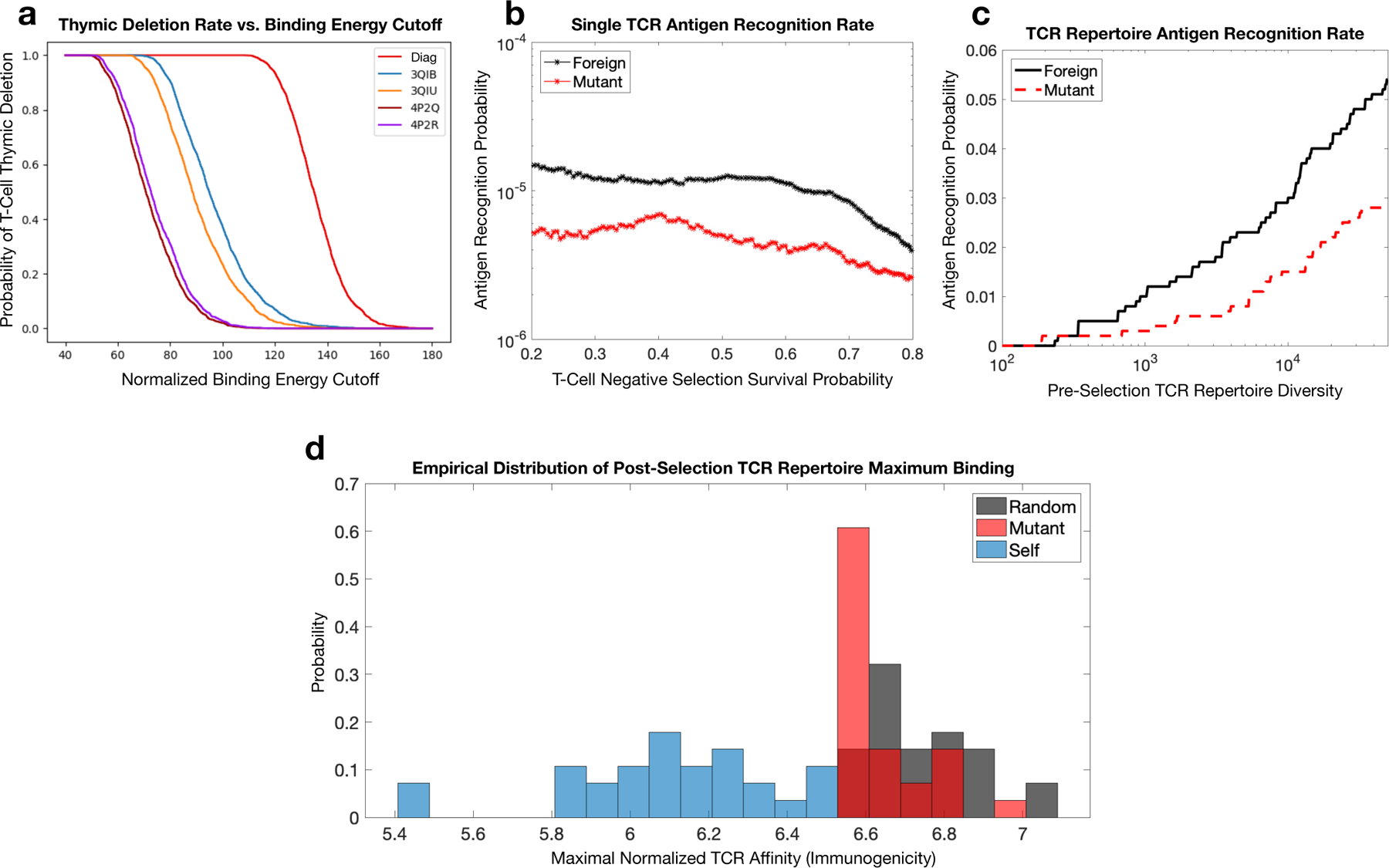
RACER-derived T-cell repertoire simulations of thymic selection and antigen recognition. a. Simulated thymic selection curves (T-cell recognition probability as a function of negative selection binding energy cutoff) incorporating the effects of non-adjacent contacts (given in Fig. 4) using Nn = 104 uniformly randomized self-peptides and Nt = 2000 randomized IEk-restricted TCRs. 4P2Q and 4P2R (purple) use T-cells generated by randomizing the CDR3 region of TCR 5cc7, while 3QIB (blue) randomizes the CDR3 of TCR 2B4, and 3QIU (yellow) randomizes the CDR3 of TCR 226 (in all cases, randomized CDR3 lengths were unchanged from the original TCR) (red curve uses RACER energy using a diagonal contact map model whose study here is motivated by previous work [12]). b. Utilizing RACER-derived energy assessments from the 2B4 crystal structure, the probability of recognizing foreign and point-mutant antigens for individual post-selection T-cells is plotted as a function of the percentage of TCRs surviving negative selection (ordinate of the graph in panel a, simulations averaged over all post-selection TCRs with pairwise interactions amongst 103 random peptides and 103 point-mutant peptides). c. The recognition probability of foreign (black) and mutant (red) peptides by the entirety of the TCR repertoire is plotted as a function of pre-selection TCR repertoire diversity (the number of unique post-selection TCRs), with negative selection thresholds giving 50% survival. d. RACER-derived immunogenicity of foreign, mutant, and self antigen. The distribution of maximum binding affinity over all post-selection T-cells for immunogenic random (gray) and point-mutated self-peptides (red) is compared to that of thymic self-peptides (blue) (There were 28 point-mutated peptides that had at least one T-cell recognition event. To keep an equal number of peptides in each distribution, these were compared with the top 28 similarly ordered foreign peptides and 28 randomly chosen self-peptide groups).
