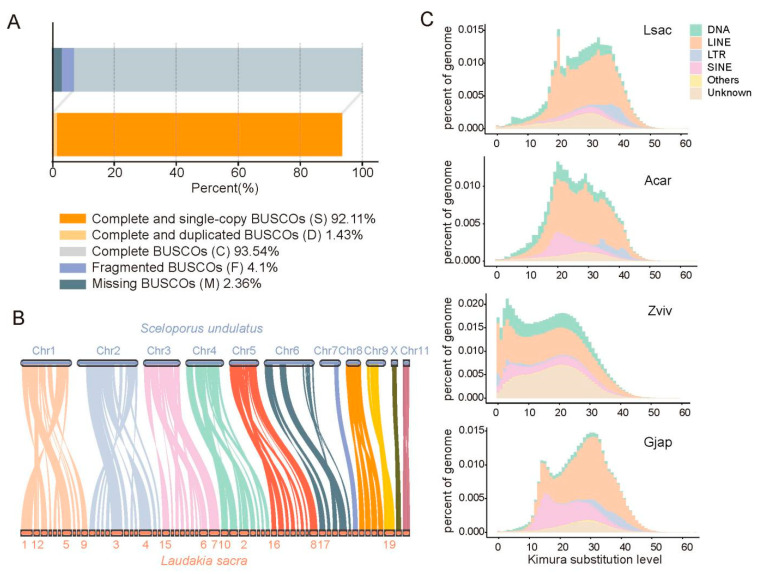
Figure 2.
Assessment of Laudakia sacra genome assembly and comparison with other lizard genomes. (A) BUSCO assessment of L. sacra assembly. (B) Genome-wide synteny between L. sacra and Sceloporus undulatus. (C) Transposable element (TE) accumulation history in different lizards. The X-axis is the CpG-adjusted Kimura distance from consensus in TE library. The Y-axis is the percentage of TE occupancy in genome. Copies clustered on right of graph markedly diverged from their corresponding consensus, potentially corresponding to ancient copies, while sequences on the left may correspond to recent copies. Lsac, L. sacra; Acar, Anolis carolinensis; Zviv, Zootoca vivipara; Gjap, Gekko japonicus.