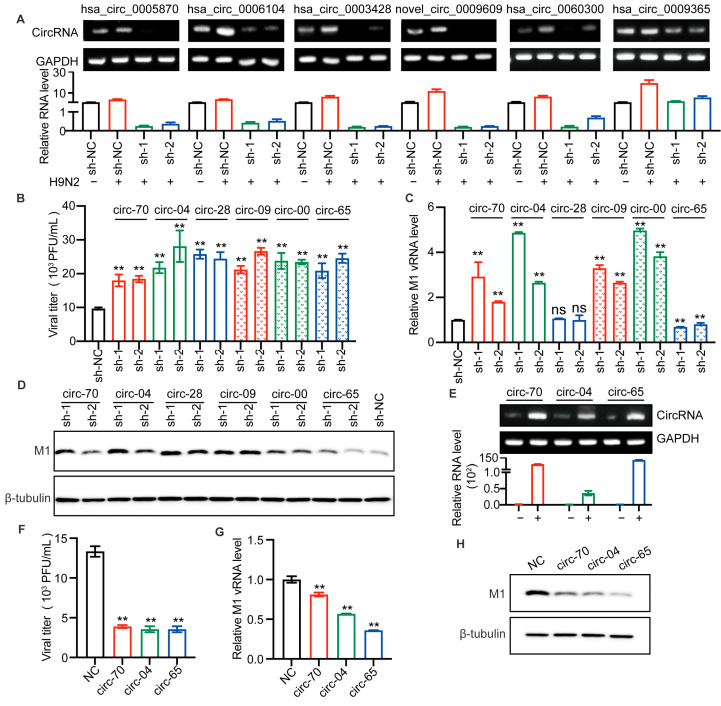
Figure 4.
Selected circRNAs affect H9N2 virus replication. (A–D) The candidate circRNAs in A549 cells were silenced by lentiviral packaged shRNAs for 48 h and then infected with H9N2 virus at 12 h (MOI = 0.5). (A) The levels of the six circRNAs were detected by RT-qPCR (n = 3) and visualized by agarose gel electrophoresis. Data were normalized to GAPDH and expressed as mean ± SD. “−” stands for PBS treatment, and “+” indicates treatment with H9N2 virus. (B) The virus titer in supernatants was measured by plaque assays, ** p < 0.01. (C) The level of AIV M1 vRNA was detected by RT-qPCR (n = 3). Data were normalized to GAPDH and expressed as mean ± SD. (D) Cell lysates were harvested for Western blotting assays using the indicated antibodies. (E–H) Overexpression of the indicated circRNAs in 293T cells by plasmid transfection for 24 h, followed by H9N2 virus infection at 12 h (MOI = 0.5). (E) The overexpression levels of the circRNAs were detected by RT-qPCR (n = 3) and visualized by agarose gel electrophoresis. Data were normalized to GAPDH and expressed as mean ± SD. “−” stands for PBS treatment, and “+” indicates treatment with H9N2 virus. Cell supernatants were harvested for virus titers (F), and for RT-qPCR (n = 3) to detect the expression level of viral M1 vRNA (G), and the remaining cell lysates were used for immunoblotting for viral M1 protein (H). Data were normalized to GAPDH and expressed as mean ± SD.