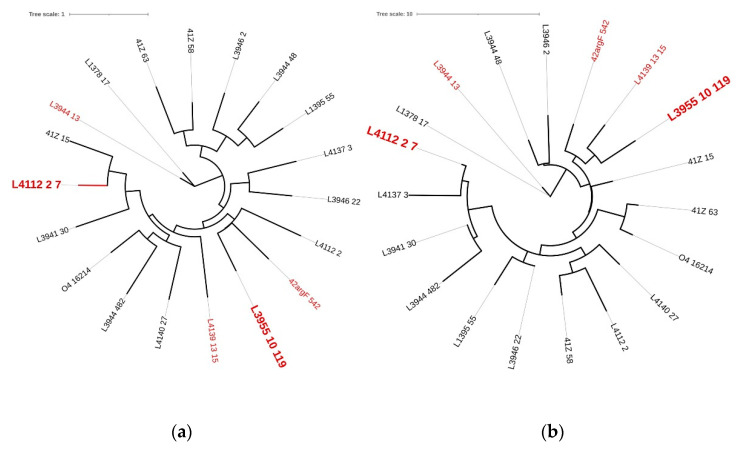
Figure 2.
Phylogenetic trees of prophage lysins based on (a) nucleotides and (b) amino acids sequences. Genomic tree was constructed using the Jukes–Cantor substitution model and no bootstrapping/likelihood as branch support method and the proteomic tree was constructed using the Le Gascuel substitution model and no bootstrapping/likelihood as branch support method in PHYML 3.3.20180621 (Geneious version 2021.1.1). Trees were visualized using iTOL v6 [30]. Red—endolysins selected for cloning; larger leaf names—selected for liposome encapsulation.