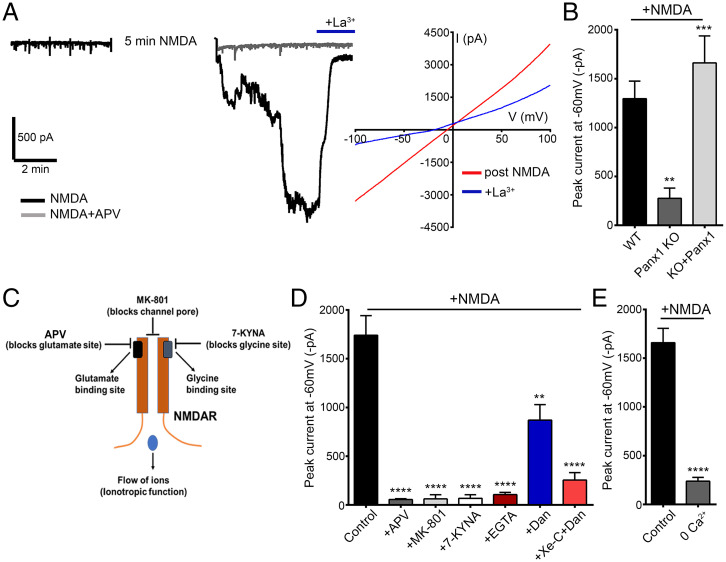
Fig. 5.
NMDAR stimulation activates downstream Panx1 currents in hippocampal neurons. (A) Representative trace (Left) from whole-cell recordings in hippocampal neurons treated for 5 min with NMDA (100 µM) in the presence or absence of APV (100 µM). The postexposure current, recorded after NMDA treatment, was inhibited by application of La3+ (100 µM). IV curves (Right), averaged from a series of recordings from NMDA-treated neurons (n = 16), before (post-NMDA) and after application of La3+. (B) Summary data for recordings in hippocampal neurons cultured from Panx1 KO (n = 12) or WT (n = 20), as well as in neurons from Panx1 KO, in which Panx1 was re-expressed (eGFP tagged; KO+Panx1 [n = 19]). **P < 0.01 and ***P < 0.001, one-way ANOVA with post hoc Bonferroni test when compared with WT or Panx1 KO, respectively. No significant difference was seen between KO+Panx1 and WT. (C) A cartoon figure illustrating binding sites for NMDAR antagonists APV (blocks glutamate binding), MK-801 (blocks channel pore and Ca2+ influx through NMDARs), or 7-KYNA (blocks glycine binding). (D) Summary of NMDA treatment–induced Panx1 currents in cultured CD1 neurons. Treatment-induced Panx1 currents were blocked when NMDA was applied in the presence of APV (100 µM, n = 9), MK-801 (20 µM, n = 8), 7-KYNA (10 µM, n = 9), or EGTA (11 mM in intracellular solution, n = 6) and reduced in the presence of dantrolene (Dan) alone (50 µM, n = 8) or Dan (10 µM) with xestospongin-C (Xe-C) (2.5 µM, n = 11). ****P < 0.0001 and **P < 0.01, one-way ANOVA with post hoc Bonferroni test when compared with control (n = 21). (E) Summary data from a series of comparable recordings show that Panx1 activation is reduced when NMDA is applied in solution containing 0 Ca2+ (n = 8) when compared with control (n = 8). For this set of recordings, NMDA was applied for a duration of 10 min. ****P < 0.0001, unpaired t test. Data are represented as mean ± SEM (B, D, and E).