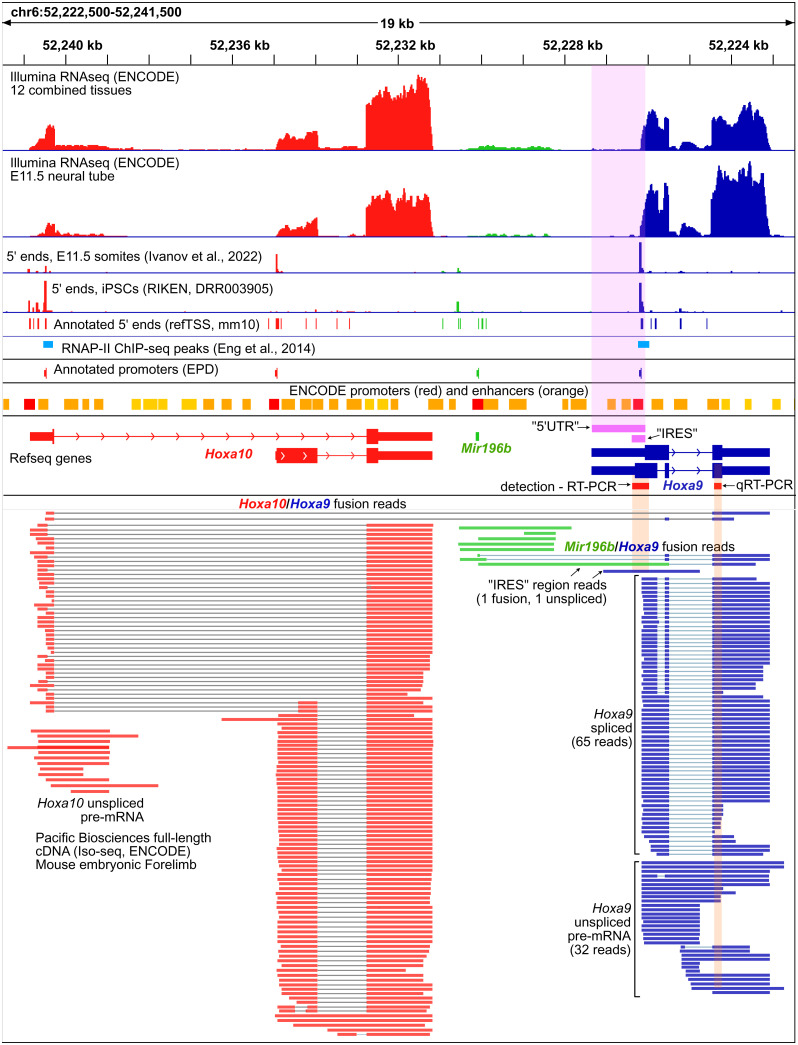
Fig. 2.
The putative extended 5′ UTR and IRES regions of mouse Hoxa9 are not expressed at biologically meaningful levels. A genome browser view showing Refseq annotations of the mouse Hoxa10/Mir196b/Hoxa9 in red, green, and blue, respectively, is shown. The putative TL(5′ UTR) and IRES regions are shown in pink. Promoters from the EPD and the ENCODE project are shown above the Refseq gene models. Illumina short-read (Upper) and PacBio full-length (Lower) RNA-seq data from the ENCODE consortium show negligible levels of RNA over the putative 5′ UTR and IRES regions. Similarly, two 5′ CAGE-seq studies [Ivanov et al. (56) and Abugessaisa et al. (53)] show no TSSs at the putative 5′ UTR, and a strong TSS peak downstream of the putative IRES. ChIP-seq peaks show RNAPII is found immediately downstream of EPD Hoxa9 and Hoxa10 promoters in mouse embryonic forelimbs [Eng et al. (29)]. PacBio RNA-seq detects Hoxa9/a10 and Hoxa9/Mir196b fusion transcripts. Regions corresponding to the PCR amplicons used previously to detect (RT-PCR) the putative Hoxa9 IRES and quantify (RT-qPCR) Hoxa9 mRNA are shown in red. The Hoxa9 RT-qPCR amplicon used for expression and polysome analysis is not specific to spliced Hoxa9 mRNA, and can amplify fusion transcripts, unspliced transcripts, and truncated transcripts initiating at refTSS annotated start sites within Hoxa9 introns.