Significance
Clear cell renal cell carcinoma (ccRCC) is a lethal cancer that is typically resistant to radiotherapy or chemotherapy. Previous research reported that Zinc Fingers And Homeoboxes 2 (ZHX2) is an oncogenic driver in ccRCC by acting as a pVHL E3 ligase substrate. In this study, we identify that USP13 is a deubiquitinase that contributes to ZHX2 stabilization in ccRCC. USP13 loss leads to decreased ccRCC cell proliferation in vitro and tumor growth in vivo. Our results suggest that USP13 is a potential therapeutic target in ccRCC by regulating ZHX2.
Keywords: ZHX2, USP13, deubiquitination, ccRCC
Abstract
Clear cell renal cell carcinoma (ccRCC) is characterized by the loss of tumor suppressor Von Hippel Lindau (VHL) function. VHL is the component of an E3 ligase complex that promotes the ubiquitination and degradation of hypoxia inducible factor α (HIF-α) (including HIF1α and HIF2α) and Zinc Fingers And Homeoboxes 2 (ZHX2). Our recent research showed that ZHX2 contributed to ccRCC tumorigenesis in a HIF-independent manner. However, it is still unknown whether ZHX2 could be modified through deubiquitination even in the absence of pVHL. Here, we performed a deubiquitinase (DUB) complementary DNA (cDNA) library binding screen and identified USP13 as a DUB that bound ZHX2 and promoted ZHX2 deubiquitination. As a result, USP13 promoted ZHX2 protein stability in an enzymatically dependent manner, and depletion of USP13 led to ZHX2 down-regulation in ccRCC. Functionally, USP13 depletion led to decreased cell proliferation measured by two-dimensional (2D) colony formation and three-dimensional (3D) anchorage-independent growth. Furthermore, USP13 was essential for ccRCC tumor growth in vivo, and the effect was partially mediated by its regulation on ZHX2. Our findings support that USP13 may be a key effector in ccRCC tumorigenesis.
In the past decades, renal cell carcinoma (RCC) incidence has been increasing steadily, although the reasons for this remain unknown. The von Hippel-Lindau (VHL) is the most frequently mutated gene in RCC, accounting for ∼80% of RCC samples (1). Numerous studies have revealed that pVHL could act as an E3 ligase component to regulate the expression of several substrates including hypoxia inducible factor α (HIF-α), Zinc Fingers And Homeoboxes 2 (ZHX2), and Scm-like with four malignant brain tumor domains 1 (SFMBT1) (1–5). Among them, the VHL-HIF pathway is one of the most characterized mechanisms contributing to clear cell RCC (ccRCC) tumorigenesis, primarily by activating the expression of multiple target genes, such as VEGF, GLUT1, and EPO (6). One of key HIF subunits, HIF2α, has been confirmed to promote the occurrence and development of kidney cancer in vitro and in vivo (1, 7, 8). Therefore, targeting HIF2α represents one of most important therapeutic strategies for treating RCC (9). On the other hand, recently developed HIF2α inhibitors could only inhibit tumor growth in certain preclinical kidney cancer models but not others (10, 11). Therefore, it may be important to target factors in HIF2α-independent signaling pathways in ccRCC, including ZHX2.
Human zinc-fingers and homeoboxes 1 (ZHX1) was identified as a protein that interacts with the transcription factor NF-YA (12). ZHX1 contains two Cys2-His2 zinc fingers and five homeodomains. A highly related protein, ZHX2, was identified and reported to function as a transcriptional repressor (13). ZHX2 can act as a tumor suppressor in hepatocellular carcinoma (HCC) by inhibiting the expression of multiple genes including Cyclin A and E and multidrug resistance1 (MDR1) expression (14, 15). On the other hand, from tissue microarray and clinicopathological analyses, ZHX2 protein expression in metastatic HCC was twice as high as in the primary lesions, indicating that ZHX2 expression is associated with metastasis in HCC (16). Recently, our research and others have established the oncogenic role of ZHX2 in various cancers, including ccRCC, breast cancer, multiple myeloma, and gastric cancer (4, 17–21). More interestingly, ZHX2 is localized on the 8q24 locus, where Myc is localized, and this locus has been reported to be amplified in many cancers, including breast cancer (22). Therefore, it is important to characterize the oncogenic role of ZHX2 in cancer, especially in ccRCC given its role as a pVHL substrate.
Deubiquitinase (DUB) and E3 ligase constitute important components of the protein ubiquitination degradation pathway. Among them, the main function of DUB is to deubiquitinate the substrate protein, thereby stabilizing the expression of the protein. At present, we have found that ZHX2 can be regulated by pVHL ubiquitination, but it remains unclear whether ZHX2 can be modified by DUB. Inhibitors of the corresponding DUB of the proto-oncoprotein could be designed in clinical applications, thereby promoting the ubiquitination and degradation of the proto-oncoprotein. In this study, we tried to find the DUB of ZHX2 that may be responsible for contributing to its protein stability regulation. Our results reveal that USP13 might be a key effector and a potential therapeutic target in ccRCC tumorigenesis by stabilizing ZHX2.
Results
Identification of USP13 as a Potential DUB for ZHX2 in ccRCC.
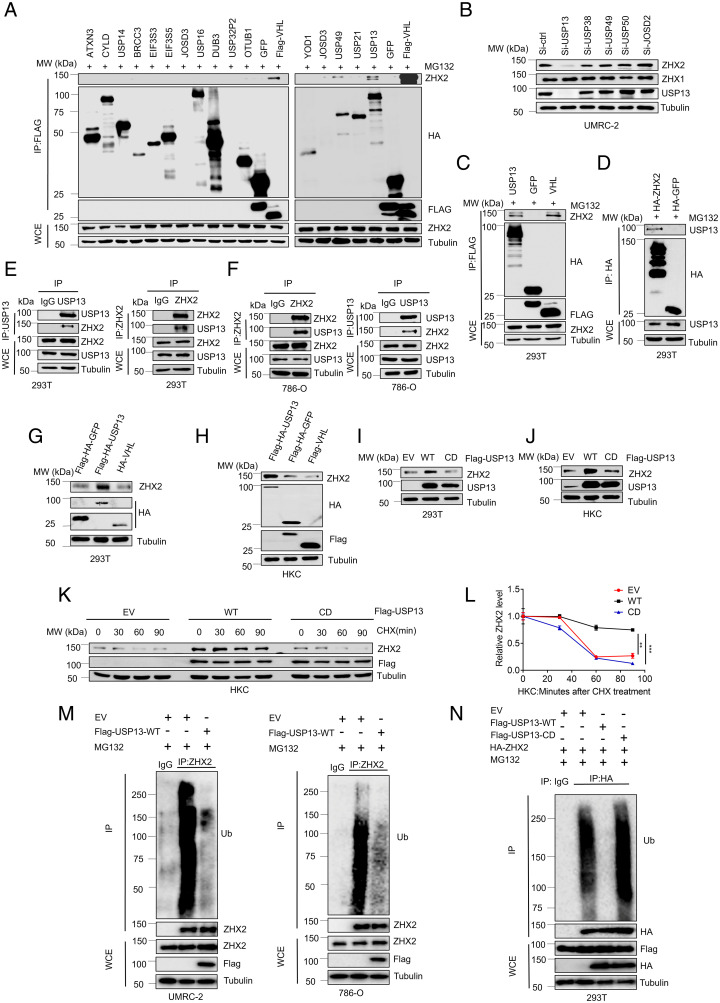
In an effort to identify the potential DUBs that regulate ZHX2, we used a previously described FLAG and hemagglutinin (HA) double-tagged DUB complementary DNA (cDNA) library (23) by transfecting them into 293T cells followed by the coimmunoprecipitation for exogenously expressed DUBs with endogenous ZHX2. In addition, we also introduced FLAG-tagged VHL, a known binding partner for ZHX2, as a positive control for our binding screening (Fig. 1A). Our binding screen identified several DUBs that showed binding with endogenous ZHX2, including USP13, USP38, USP49, USP50, and JOSD2 (Fig. 1A and SI Appendix, Fig. S1 A–E). As these are short-term transfection experiments, it is important to note that the expression levels of each individual DUB may be different. This differential DUB expression may limit the number of positive binding partners for ZHX2 in our screening. Nonetheless, we performed follow-up experiments for these identified ZHX2 binding partners with a smart-pool small interfering RNA (siRNA) against each member, in which each siRNA pool down-regulated the corresponding gene efficiently (SI Appendix, Fig. S1F). Among them, we found that depletion of USP13, but not the other DUBs, down-regulated ZHX2 protein levels in ccRCC cell lines (UMRC-2) (Fig. 1B). It is important to note that none of these siRNA transfections led to a change in ZHX1 protein levels, suggesting that the effect of USP13 on ZHX2 is specific (Fig. 1B). Next, we validated semiendogenous interaction between ZHX2 and USP13 by using reciprocal immunoprecipitations (Fig. 1 C and D). Motivated by these findings, we also performed endogenous immunoprecipitation between ZHX2 and USP13 in either 293T cells or the ccRCC cell line 786-O. With reciprocal immunoprecipitations, we confirmed the endogenous binding between ZHX2 and USP13 in all of these cell lines (Fig. 1 E and F).
Fig. 1.
USP13 interacts with ZHX2 and promotes USP13 protein stability. (A) Representative immunoblots of immunoprecipitations (IPs) and whole-cell extracts (WCEs) from 293T cells transfected with FLAG and HA double-tagged DUB cDNA library (4 µg plasmid per p60 plate) as indicated and then treated with 10 μM MG132; FLAG-HA-GFP is the negative control, and FLAG-VHL is the positive control. (B) Immunoblots of the lysates from UMRC-2 transfected with siRNAs of some candidate DUB proteins. (C and D) Immunoblots of the WCE and IP from 293T transfected with FLAG-HA-USP13, FLAG-HA-GFP, FLAG-VHL, HA-ZHX2, and HA-GFP as indicated. (E and F) Immunoblots of the WCE and IP from 293T and 786-O cells as indicated. (G and H) Immunoblots of the lysates from 293T and HKC transfected with FLAG-HA USP13, FLAG-HA-GFP, and FLAG/HA-VHL as indicated. (I and J) Immunoblots of lysates from 293T and HKC cells transfected with EV, WT, or CD form of USP13 as indicated. (K) Immunoblots of lysates from HKC cells transfected with EV, WT, or CD form of USP13 and then treated with 10 μg/mL CHX for the indicated time. (L) Quantification of ZHX2 expression level shown. (M) Immunoblots of WCE or IP from UMRC-2 or 786-O cells infected with FLAG-USP13 or empty vectors and then treated with 10 μM MG132 as indicated. (N) Immunoblots of WCE or IP from 293T cells transfected with indicated plasmids and then treated with 10 μM MG132 as indicated. Error bars, SEM; **P < 0.01; ***P < 0.001; MW, molecular weight.
USP13 is a DUB that was previously reported to be involved in the deubiquitination of various substrates, including MCL1, SKP2, MITF, PTEN, TOPBP1, and c-MYC (24–29). Additionally, USP13 was reported to regulate SIAH2 ligase stability via noncatalytic ubiquitin-binding domains (30). Given that USP13 and ZHX2 can interact, we aimed to elucidate the role of USP13 on regulating ZHX2 protein stability. First, we overexpressed USP13 in 293T cells and found that USP13 led to increased ZHX2 protein levels in 293T cells or HKC cells while VHL overexpression caused down-regulation of ZHX2 (Fig. 1 G and H), reinforcing the role of pVHL as the E3 ligase for ZHX2 as shown previously (4). To confirm if the expression of ZHX2 is dependent on the enzymatic activity of USP13, we transfected HKC and 293T cell lines with USP13-wild type (WT) or the catalytically inactive USP13 (CD) plasmid (28). WT, but not catalytically inactive USP13 (CD), up-regulated ZHX2 protein levels (Fig.1 I and J), revealing that USP13 overexpression leads to the up-regulation of the ZHX2 protein level in an enzymatically dependent way. Next, we tried to examine whether USP13 can act as a DUB for ZHX2 and promote ZHX2 deubiquitination and protein stability in ccRCC cells. To explore this, we applied the protein synthesis inhibitor (cycloheximide [CHX]) to treat an HKC cell line expressing EV, USP13-WT, or USP13-CD and detected ZHX2 protein expression at indicated times. Overexpression of USP13-WT displayed increased ZHX2 protein stability by showing a slower protein degradation rate compared to either USP13-CD or EV groups (Fig. 1 K and L). Furthermore, we also examined ZHX2 protein ubiquitination in cells upon USP13 overexpression. Under denaturing conditions, we found that USP13 WT overexpression led to decreased ZHX2 ubiquitination in ccRCC cell lines (UMRC-2 and 786-O) (Fig. 1M). In accordance with these findings, we also found that WT, and not CD, USP13 promoted ZHX2 deubiquitination in 293T cells (Fig. 1N). In summary, our data reveal that USP13 serves as a previously uncharacterized ZHX2 DUB in ccRCC.
USP13 Depletion Leads to Decreased ZHX2 Stability in ccRCC.
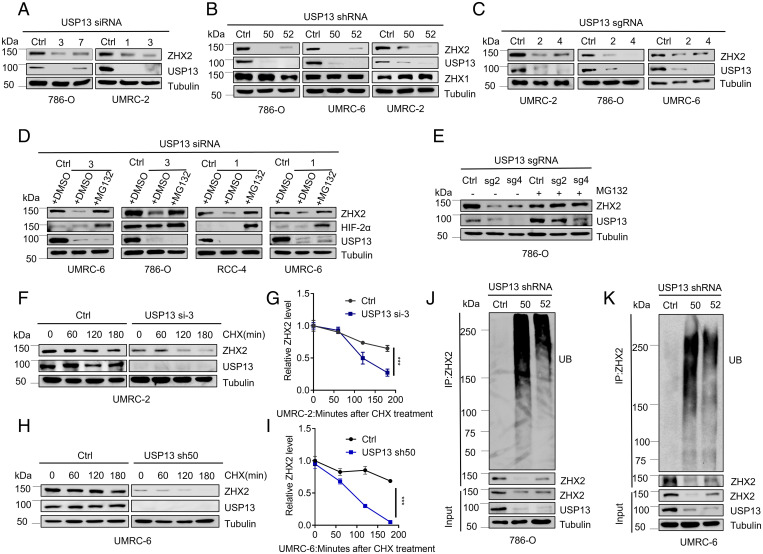
To test whether USP13 is critical for ZHX2 regulation in kidney cancer cell lines, we transfected several independent USP13 siRNAs in several ccRCC cell lines, including UMRC-2 and 786-O cells. USP13 depletion led to ZHX2 protein level down-regulation in cells above (Fig. 2A). It is important to note that the loss of USP13 did not consistently affect ZHX2 messenger RNA (mRNA) levels in several different cell lines (SI Appendix, Fig. S2A). Additionally, we also obtained two independent USP13 short hairpin RNAs (short hairpin RNAs) (#50 and #52) and found that USP13 short hairpin RNA-mediated depletion also led to consistent ZHX2 protein down-regulation in multiple ccRCC cell lines, including 786-O, UMRC-6, and UMRC-2 (Fig. 2B). To further confirm this, we constructed multiple USP13-single guide RNAs (sgRNAs) by CRISPR-Cas9. Our single guide RNAs (#2 and #4) efficiently decreased USP13 protein expression in several ccRCC cells (UMRC-2, RCC4, 786-O, and UMRC-6), which corresponded with a decreased expression of ZHX2, strengthening the regulation of USP13 on ZHX2 (Fig. 2C and SI Appendix, Fig. S2B).
Fig. 2.
Loss of USP13 destabilizes ZHX2 by promoting ZHX2 ubiquitination in kidney cancer cells. (A–C) Immunoblots of lysates from indicated cells transfected with siRNA (A) targeting USP13 or from cells infected with lentivirus encoding short hairpin RNA (B) or single guide RNAs (C) targeting USP13. (D and E) Immunoblots of lysates from UMRC-6, 786-O, RCC-4, and UMRC-6 cells transfected with siRNAs (#1 or #3) targeting USP13 (D) or from 786-O infected with USP13 single guide RNAs (sg2 or sg4) (E), and then treated with DMSO or MG132 as indicated. (F–I) Immunoblots of lysates from UMRC-2 (F) or UMRC-6 (I) cells infected with either Ctrl siRNA, USP13 si-3, Ctrl short hairpin RNA, and USP13 sh50 and then treated with 10 μg/mL CHX for the indicated time. G and I are the quantification of the ZHX2 expression level shown in F and H. (J and K) Immunoblots of WCEs and IPs (endogenous ubiquitination level of ZHX2, under denaturing conditions) from 786-O and UMRC-6 cells infected with lentivirus encoding either Ctrl short hairpin RNA targeting USP13. Error bars, SEM; ***P < 0.001.
To further explore whether depletion of USP13 could regulate ZHX2 stability in ccRCC cell lines, we treated these cells with a proteasomal inhibitor, MG132, and found that USP13 depletion–mediated ZHX2 down-regulation could be rescued by MG132 in several ccRCC cell lines (Fig. 2D and SI Appendix, Fig. S2C), suggesting that USP13 mainly affects ZHX2 protein stability. Accordingly, USP13 depletion by single guide RNAs down-regulated ZHX2 protein levels in 786-O, the phenotype ameliorated by concurrent treatment with the proteasomal inhibitor MG132 (Fig. 2E). Furthermore, CHX experiments revealed that the depletion of USP13 by either siRNA or short hairpin RNA could significantly facilitate the protein degradation rate of ZHX2 in UMRC-2 and UMRC-6 cells, respectively (Fig. 2 F–I). In addition, we also examined ZHX2 ubiquitination levels in UMRC-6 and 786-O cell lines. In these two cells, USP13 depletion by two USP13 short hairpin RNA increased the ZHX2 ubiquitination level (Fig. 2 J and K).
To further examine the ubiquitination sites of ZHX2 regulated by USP13, we searched the phosphosite database (https://www.phosphosite.org/homeAction.action) and retrieved four potential ZHX2 ubiquitination sites (K455, K542, K599, and K664). We mutated these potential sites individually to alanine (K-A). Upon USP13 depletion, we observed increased WT ZHX2 ubiquitination, consistent with our data shown in the paper (Fig. 2 J and K). K455A, K599A, or K664A (but not K542A) all led to diminished ZHX2 ubiquitination compared to WT upon USP13 depletion in 293T cells (SI Appendix, Fig. S3), suggesting they are potential ZHX2 ubiquitination sites regulated by USP13. Future research needs to be further performed to pinpoint the role of these sites on regulating ZHX2 protein stability. In conclusion, our results suggest that USP13 depletion down-regulated the ZHX2 level by increasing ZHX2 ubiquitination and degradation. Thus, USP13 serves as a bona fide DUB for ZHX2 in ccRCC.
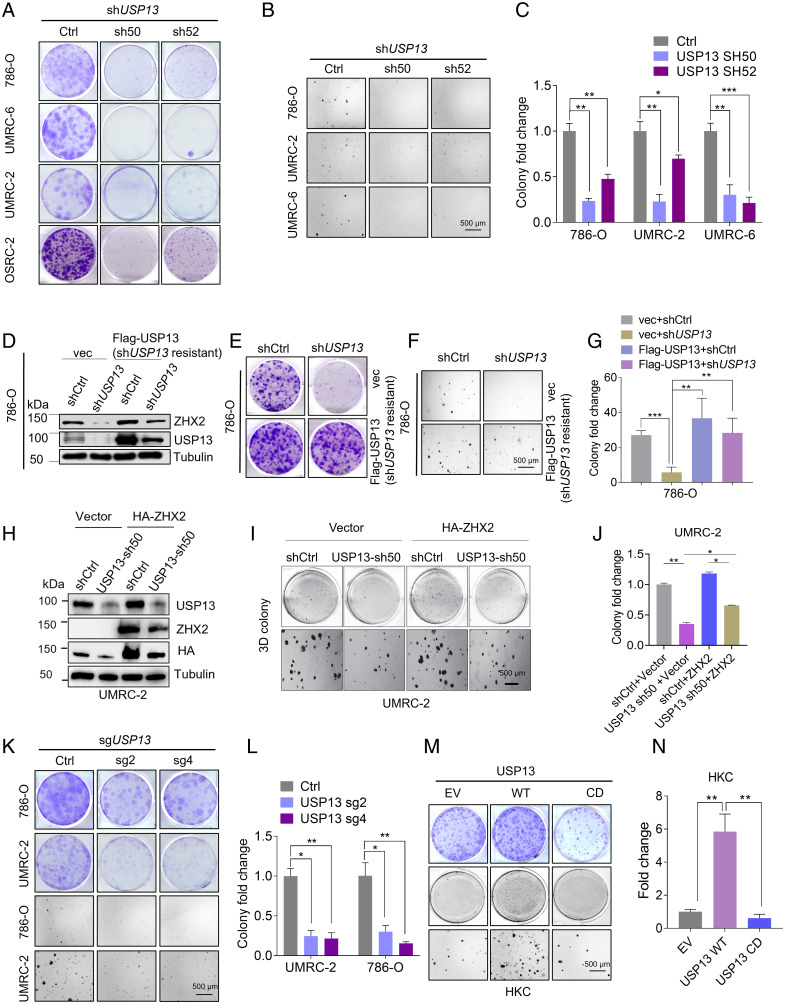
USP13 Is Essential for Two-Dimensional (2D) and Three-Dimensional (3D) ccRCC Cell Growth.
Given that USP13 is a critical regulator in ZHX2 protein stability and ZHX2 serves as an oncoprotein in kidney cancer (4), we aimed to interrogate the function of USP13 depletion in kidney cancer. Firstly, we conducted 2D colony growth assays and MTS assays in multiple ccRCC cell lines (OSRC-2, 786-O, UMRC-6, and UMRC-2). Our results showed that depletion of USP13 by two independent short hairpin RNAs inhibited colony formation and cell proliferation (SI Appendix, Fig. S4 A and B and Fig. 3A). Furthermore, 3D soft agar experiments were performed in cell lines described above, and our results also showed that depletion of USP13 inhibited soft agar growth (Fig. 3 B and C). Next, to explore whether the effect of USP13 short hairpin RNA is on-target, we infected 786-O with the mutant USP13 cDNA that was resistant to USP13 short hairpin RNA with silent mutations in the short hairpin RNA recognition region. Our results showed that USP13 short hairpin RNA inhibited ZHX2 expression and ccRCC cell growth, while these changes were rescued by the USP13 cDNA (short hairpin RNAs resistant) (SI Appendix, Fig. S4C and Fig. 3 D–G), revealing that the effect of USP13 short hairpin RNA on ZHX2 and soft agar growth was on-target. To determine whether the effect of USP13 on ccRCC growth is mediated through its regulation on ZHX2, we also overexpressed ZHX2 in 786-O and UMRC-2 cells following USP13 depletion (USP13 sh50 or sh52) (SI Appendix, Fig. S4 D and G and Fig. 3H). Our 2D colony growth assays or soft agar growth assays showed that USP13 short hairpin RNA led to decreased cell proliferation, the phenotype ameliorated at least partially by ZHX2 overexpression in these cells (SI Appendix, Fig. S4 D–I and Fig. 3 H–J). To further strengthen our findings on the function of USP13 depletion in ccRCC cell lines, we also used USP13 single guide RNAs above to inhibit USP13 expression and thereby observed the phenotype of multiple ccRCC cell lines by 2D or 3D assays. Similarly, our results showed that depletion of USP13 by single guide RNAs also inhibited cell proliferation and colony growth (SI Appendix, Fig. S4 J and K and Fig. 3 K and L). Conversely, we also overexpressed USP13 (WT or CD) in HKC cells (normal renal epithelial cell) and found that whereas WT USP13 promoted colony formation or soft agar growth, CD USP13 failed to do so (SI Appendix, Fig. S4L and Fig. 3 M and N). To further explore the potential effect of USP13 down-regulation in HKC cells, we depleted USP13 expression by two different single guide RNAs (#2, #4). Whereas one single guide RNA (#2) could slightly inhibit HKC 2-D or 3D cell growth, the other one did not affect HKC cell growth (SI Appendix, Fig. S4 M–O). Our results suggest that compared to ccRCC cells, USP13 depletion does not lead to a robust cell proliferation defect in normal renal epithelial cells. In addition, USP13 promotes ccRCC cell proliferation in an enzymatic-dependent manner. In summary, USP13 is essential for ccRCC cell proliferation and tumorigenic potential. Furthermore, to determine how USP13 depletion leads to decreased ccRCC cell proliferation, we examined the apoptosis rate of ccRCC cells in these cells. USP13 depletion induced cell apoptosis in ccRCC cells (SI Appendix, Fig. S4 P and Q). We also examined the cell cycle distribution of ccRCC cells infected with USP13 short hairpin RNAs or control (shCtrl) but did not observe a gross difference among all groups (SI Appendix, Fig. S4 R and S). Therefore, our results suggest that USP13 knockdown affects cell growth at least partially through cell apoptosis.
Fig. 3.
USP13 is important for ccRCC tumor cell growth. (A) Representative crystal violet staining of indicated cells infected with either Ctrl short hairpin RNA or short hairpin RNA targeting USP13 (786-O, UMRC-2, UMRC-6, and OSRC-2). (B and C) Representative 3D soft agar growth pictures (B) and quantification of colony numbers (triplicate wells) (C) of 786-O, UMRC-2, or UMRC-6 cells infected with either Ctrl short hairpin RNA or short hairpin RNA targeting USP13. (D and E) Immunoblots of lysates (D) and crystal violet staining (E) of 786-O cells infected with lentivirus encoding shCtrl, shUSP13 (sh50) or pLenti6-vec, pLenti6-FLAG-USP13 (shUSP13-resistant) as indicated. (F and G) Representative 3D soft agar growth pictures (F) and quantification of colony numbers (triplicate wells) (G) of 786-O cells infected with lentivirus encoding shCtrl, shUSP13 (sh50) or pLenti6-vec, pLenti6-FLAG-USP13 (shUSP13-resistant) as indicated. (H–J) Immunoblots of lysates (H), representative 3D soft agar growth pictures (I), and quantification of colony numbers (triplicate wells) (J) of UMRC-2 cells infected with lentivirus encoding Ctrl, USP13 sh50 or pLenti6-vec, pLenti6-HA-ZHX2 as indicated. (K and L) Representative crystal violet staining (K), 3D soft agar growth pictures (K), and quantification of indicated cells infected with either Ctrl single guide RNA or single guide RNA targeting USP13 (786-O and UMRC-2) (L). (M and N) Representative crystal violet staining (J) or representative 3D soft agar growth pictures (M) and quantification of colony numbers (triplicate wells) (N) of indicated cells infected with EV, WT, or CD form of USP13 as indicated. Error bars, SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
USP13 Is Critical for ccRCC Tumor Formation In Vivo.
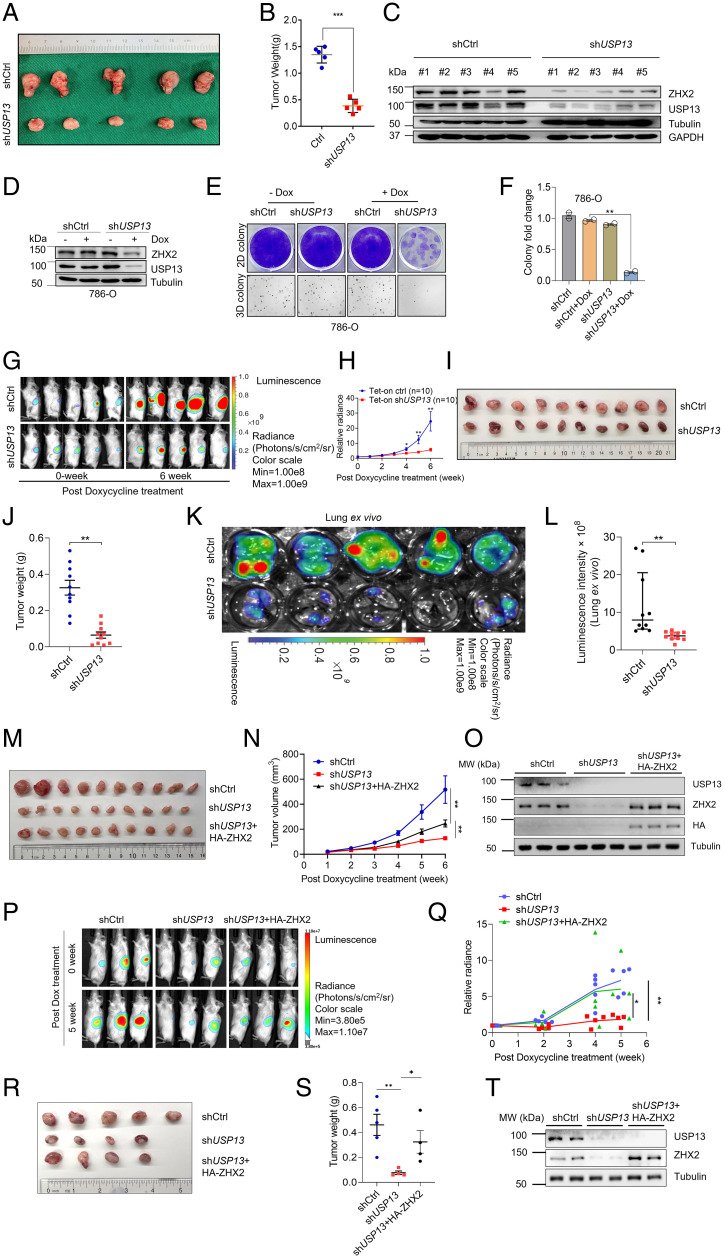
Motivated by our in vitro phenotype experiments with USP13 knockdown or overexpression, next we aimed to determine the effect of USP13 on ccRCC tumor growth in vivo. First, we injected previously established USP13 knockdown ccRCC cells (sh50, Fig. 3A) subcutaneously into BALB/C nude mice followed by monitoring tumor growth over time. We found that USP13 depletion led to decreased tumor volume and tumor weight (Fig. 4 A and B). We also extracted cell lysates from these tumors upon necropsy to examine whether this effect correlated with USP13 and ZHX2 knockdown. We found that tumors derived from USP13-depleted ccRCC cells displayed decreased USP13 and concordantly lower ZHX2 levels, arguing that the decreased tumor growth in vivo was mostly likely due to the efficient knockdown of USP13 and decreased ZHX2 in ccRCC (Fig. 4C).
Fig. 4.
Depletion of USP13 suppresses ccRCC tumorigenesis. (A–C) Image of tumors (A), tumor weights (B), and immunoblots of the lysates from tumors (C) of OSRC-2 cell infected with lentivirus encoding control (shCtrl) or USP13 short hairpin RNA (sh50) and injected subcutaneously at the armpit of BALB/C nude mice. (D–F) Immunoblots of lysates (D); crystal violet staining, 3D soft agar growth pictures (E); and quantification of colony numbers (triplicate wells) (F) of 786-O infected with either doxycycline-inducible Ctrl short hairpin RNA or doxycycline-inducible short hairpin RNA targeting USP13 (sh50) and then treated with doxycycline as indicated. (G–J) Representative imaging of before (0 wk) and 6 wk post–doxycycline treatment (G), quantification of post–doxycycline treatment bioluminescence imaging (H), image of tumors (I), and tumor weight (J) from 786-O luciferase stable cells infected with lentivirus encoding either doxycycline-inducible Ctrl short hairpin RNA (Tet-on ctrl) or doxycycline-inducible short hairpin RNA targeting USP13 (Tet-on shUSP13) and injected orthotopically into the renal capsule of NSG mice. Two-way ANOVA analysis was performed for H and J. (K and L) Representative lung ex vivo bioluminescence imaging (K) and quantification of ex vivo imaging from Tet-on Ctrl and Tet-on shUSP13 groups (L). (M–O) Quantification of post–doxycycline treatment tumor volume (M), image of tumors (N), and immunoblots of the lysates from tumors (O) of 786-O infected with either doxycycline-inducible Ctrl short hairpin RNA, doxycycline-inducible short hairpin RNA targeting USP13 (sh50), or doxycycline-inducible short hairpin RNA targeting USP13 with HA-ZHX2; injected subcutaneously at the armpit of BALB/C nude mice; and then treated with doxycycline as indicated. (P–T) Representative imaging of before (0 wk) and 6 wk post–doxycycline treatment (P), quantification of post–doxycycline treatment bioluminescence imaging (Q), image of tumors (R), tumor weight (S), and immunoblots of the lysates from tumors (T) of 786-O infected with either doxycycline-inducible Ctrl short hairpin RNA, doxycycline-inducible short hairpin RNA targeting USP13 (sh50), or doxycycline-inducible short hairpin RNA targeting USP13 with HA-ZHX2; then treated with doxycycline as indicated; and injected orthotopically into the renal capsule of NSG mice. Two-way ANOVA analysis was performed for Q and S. Error bars, SEM; *P < 0.05; **P < 0.01; ***P < 0.001. Dox, doxycycline.
Next, to examine whether USP13 may be important for ccRCC tumor growth maintenance, we infected 786-O with USP13 short hairpin RNA (doxycycline inducible) and observed a decreased USP13 expression after treating cells with doxycycline (Fig. 4D), which is consistent with decreased colony formation on both 2D and 3D (Fig. 4 E and F). Then, we infected these cells with firefly luciferase vectors and injected these cells orthotopically into kidney capsules of NSG mice. Upon tumor growth establishment, we fed the mice with doxycycline chow, and bioluminescence imaging was conducted to monitor the tumor growth rate of ccRCC cells. USP13 depletion led to decreased tumor growth in kidney that was monitored by bioluminescence imaging (Fig. 4 G and H). At necropsy of mice, USP13 depletion in vivo displayed reduced tumor burden compared to control mice (Fig. 4 I and J). We also detected the bioluminescence intensity in lung to examine whether USP13 could affect the lung metastasis of ccRCC in vivo and found that depletion of USP13 inhibited lung metastasis (Fig. 4 K and L). In summary, USP13 is critical for ccRCC tumorigenesis in vivo.
To examine whether ZHX2 overexpression would rescue the phenotype of USP13 depletion in vivo, we have performed subcutaneous or kidney orthotopic xenograft experiments with rescue experiments mentioned above. First, we injected previously established doxycycline-inducible USP13 short hairpin RNA 786-O cells (Fig. 4D) or USP13 short hairpin RNA 786-O cells infected with HA-ZHX2 subcutaneously into immunodeficient NSG mice followed by tumor growth monitoring over time. Our results showed that ZHX2 overexpression could at least partially rescue decreased tumor volume caused by USP13 depletion (Fig. 4 M and N). Upon necropsy, we extracted cell lysates from these tumors and confirmed the expression of ZHX2 and USP13 in these tumors, suggesting that the partial tumor growth rescue was due to ZHX2 overexpression (Fig. 4O). To further verify our findings, we also injected these cells orthotopically into the kidney capsules of NSG mice. Consistent with the subcutaneous xenograft experiments, ZHX2 overexpression at least partially rescued the tumor growth defect phenotype in 786-O cells with USP13 depletion (Fig. 4 P–S), which was also further confirmed by Western blots (WBs) in these tumors upon necropsy (Fig. 4T). In summary, our data suggest that ZHX2 is one of the key mediators for the effect of USP13 in ccRCC.
USP13 Regulates Gene Expression Involved in ZHX2 Signaling in ccRCC.
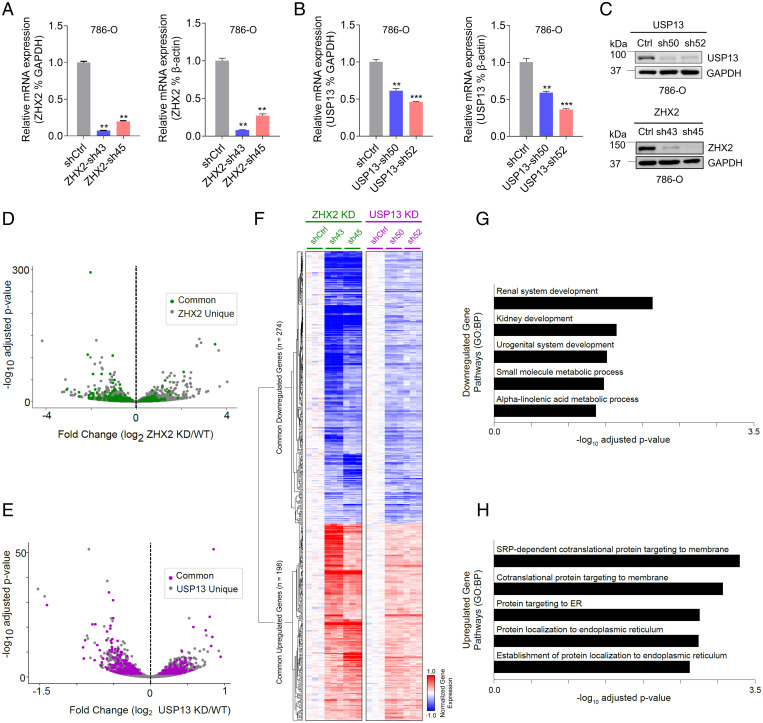
Considering that USP13 may regulate ZHX2 protein stability and therefore control ccRCC tumorigenesis, it is reasonable to postulate that USP13 and ZHX2 perturbation may impact the expression of some common genes. To uncover the potential common downstream pathways that may be regulated by both USP13 and ZHX2, we decided to generate ccRCC cell lines with ZHX2 or USP13 knockdown. First, we generated 786-O cells with two independent ZHX2 short hairpin RNAs (sh43, sh45), and RT-PCR results confirmed efficient ZHX2 depletion (∼80 to 90% messenger RNA depletion) by using either GAPDH or β-actin as controls. We also generated 786-O cells with two independent USP13 short hairpin RNAs (sh50, sh52) (Fig. 5A). Despite repeated efforts, we managed only to get partial USP13 depletion (∼50 to 60% depletion) in USP13 short hairpin RNA-infected cells (Fig. 5B). Nonetheless, our western blot showed that we could achieve efficient USP13 down-regulation (Fig. 5C). Next, we performed RNA sequencing (RNA-seq) analyses in 786-O cells with either ZHX2 depletion or USP13 depletion. Interestingly, ZHX2 commonly regulated genes could partially overlap with USP13 regulated genes both in up-regulated and down-regulated groups (up-regulated: USP13-198/812, ZHX2-198/2146; down-regulated: USP13-274/943, ZHX2-274/2003), as indicated by volcano plots and analysis results (Fig. 5 D and E and SI Appendix, Table S1). These commonly regulated genes after either ZHX2 or USP13 depletion shared similar regulation patterns across both conditions (Fig. 5F). Our RNA sequencing data analysis strongly suggests that USP13 and ZHX2 regulated a subset of target genes important in ccRCC (Fig. 5 G and H). For example, for the down-regulated gene list upon USP13 and ZHX2 depletion, there appeared to be an enrichment for pathways involved in renal system development and kidney development (Fig. 5 G and H). We also confirmed that the expression of genes involved in these pathways was down-regulated by either USP13 or ZHX2 depletion (SI Appendix, Fig. S5A). In addition, we also noticed that some of pathways related to endoplasmic reticulum (ER) were affected by both USP13 and ZHX2 knockdown (Fig. 5H). Lipid storage was reported to be important for maintaining ER membrane homeostasis in ccRCC (31). To examine whether USP13 or ZHX2 depletion affected lipid metabolism, and therefore contributing to ER homeostasis in ccRCC, we also performed seahorse experiments to determine the oxygen consumption rate (OCR) in cells depleted of USP13 or ZHX2. Neither USP13 nor ZHX2 depletion led to any significant change in lipid-mediated OCR, suggesting that USP13 or ZHX2 may not grossly regulate lipid metabolism in ccRCC (SI Appendix, Fig. S5 B and C). It is also important to point out that due to the incomplete depletion of USP13 messenger RNA in these USP13 short hairpin RNA–infected ccRCC cells, it is likely that we might be missing some other commonly regulated genes involved in ccRCC, which awaits future investigation.
Fig. 5.
USP13 regulates gene expression involved in ZHX2 signaling in ccRCC. (A–C) Quantification of messenger RNA level (A and B) and protein level (C) of USP13 and ZHX2 in 786-0 infected with either USP13 short hairpin RNA (sh50 and sh52) or ZHX2 short hairpin RNA (sh43 and sh45) using either GAPDH or actin as controls. (D and E) Volcano plots of ZHX2 (D) or USP13 (E) commonly regulated genes. (F) A hierarchically clustered heatmap of ZHX2 and USP13 commonly regulated genes. (G and H) Pathway enrichment analysis of commonly down-regulated (G) or up-regulated (H) genes between USP13 and ZHX2. Error bars, SEM; **P < 0.01; ***P < 0.001.
To investigate whether some of previously established ZHX2 downstream target genes (such as NF-κB pathway genes) were regulated by USP13 depletion (4), we examined their expression by qRT-PCR in USP13-depleted cells. USP13 depletion by the previously characterized siRNA led to decreased expression in some NF-κB genes, such as TNF, VCAM1, and ICAM1, but not BCL2 (SI Appendix, Fig. S5D). To obtain a more comprehensive readout of ZHX2 target changes after USP13 depletion, we examined how our shUSP13 RNA sequencing data corresponded to the 390 coordinately down-regulated ZHX2 and p65 genes identified in our previous study in ccRCC (4). Among them, 17.7% (69 of 390) overlapped with USP13 knockdown down-regulated genes (SI Appendix, Fig. S5E and Table S2). This was a significantly larger overlap than expected by random chance (P < 0.0001) as tested with a permutation test against all expressed genes (random chance would assume 19.75 ± 4.31 genes; mean ± SD). Regardless, our gene expression data show that USP13 knockdown does coincide with the down-regulation of ZHX2 and subsequently affects some but not all downstream ZHX2-NF-κB target genes. In summary, our results suggest that USP13 regulates ZHX2 signaling in ccRCC.
USP13 Signaling Is Dysregulated in ccRCC Patients.
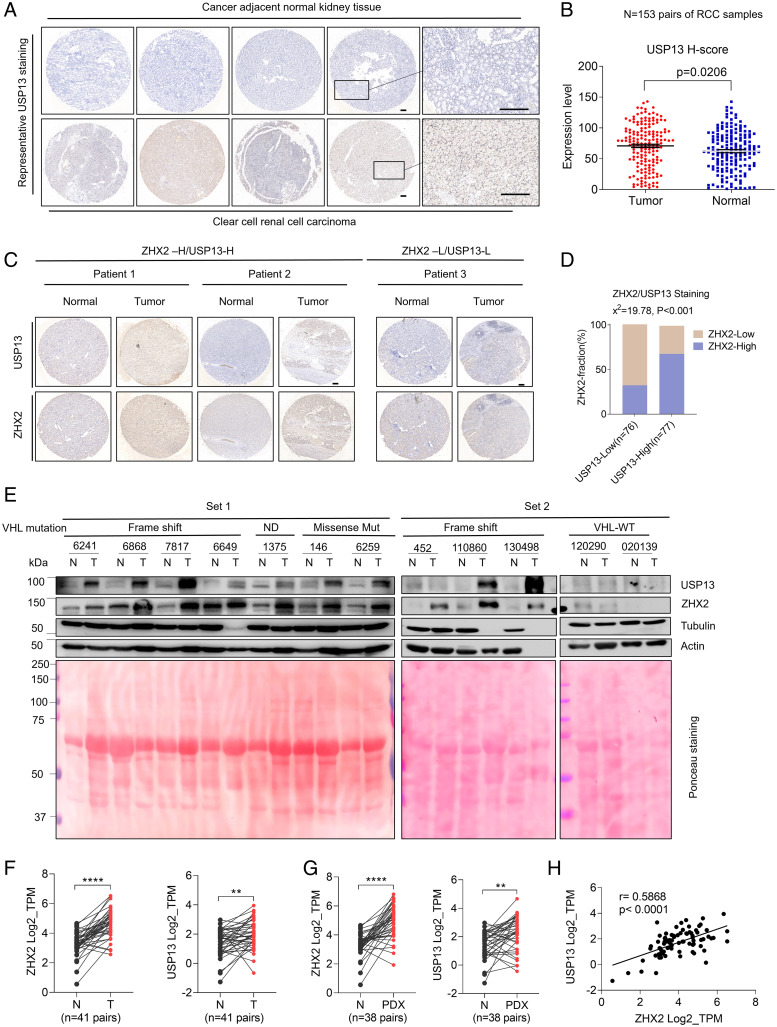
To explore the clinical relevance of USP13 and ZHX2 in ccRCC, we first performed immunohistochemistry (IHC) staining for USP13 in a previously characterized ccRCC tumor microarray set (32), which contains 153 pairs of ccRCC tumors and matched normal tissue. USP13 displayed higher expression in RCC tumors compared to normal tissue samples (Fig. 6 A and B). Next, we aimed to examine the correlation between USP13 and ZHX2 in these clinical specimens. To this end, we also stained this cohort of samples with ZHX2 and USP13 antibodies. We assigned USP13 and ZHX2 staining intensity to low or high groups defined by their H-score (median) as described previously (33, 34). We found that among USP13-low staining samples, they appeared to be weak with ZHX2 staining. Comparatively, for those samples with high USP13 staining intensities, there were significantly more samples with high ZHX2 staining intensity. Together, this suggests a significant clinical association of ZHX2-USP13 in RCC patients (P < 0.001) (Fig. 6 C and D).
Fig. 6.
USP13 level correlates with ZHX2 expression in ccRCC tumor tissues. (A and B) Representative USP13 IHC staining pictures (A) and quantification of relative USP13 signal intensity (H-SCORE) of tumor and paired normal tissues (B) from a 153-pair ccRCC cohort. Bar, 100 μm. (C and D) Representative IHC staining images (C) and correlation analysis (D) of ccRCC TMAs with two staining grades (H, high; L, low) showing the expression correlation between UPS13 and ZHX2 protein. H-SCORE was used for indicating the immunostaining intensity of the sample. According to the distribution of the H-SCORE, we used the median method to group the 153-pairs ccRCC cohort. Then we used the χ2 test to analyze the correlation between the two proteins (D) Bar, 100 μm. (E) Immunoblots of lysates from paired normal and kidney tumor tissues (two sets). VHL mutation status is as labeled. ND, not determined. Total input protein of tissues is shown by Ponceau S staining. (F and G) ZHX2 and USP13 messenger RNA levels between normal and paired ccRCC tumors (n = 41) from the gene expression database available through University of Texas Southwestern Medical Center SPORE (European Genome-Phenome Archive under the accession number EGAS00001005516) (35). (H) Correlation between ZHX2 and USP13 messenger RNA level in ccRCC tumors (n = 41) from the gene expression database available through University of Texas Southwestern Medical Center Kidney SPORE program (35).
Additionally, we also extracted cell lysates from ccRCC tumor samples and corresponding normal kidney tissues. Among 8 pairs of tissues that displayed VHL a missense mutation, a frame-shift mutation, or a splice variant as characterized previously (4), USP13 levels were up-regulated in tumors, coinciding with increased ZHX2 protein levels in these samples (Fig. 6E). However, for the tumor tissues that displayed VHL WT, neither USP13 nor ZHX2 displayed the significant up-regulation of protein levels in tumor tissues (Fig. 6E). Interestingly, USP13 displayed lower protein levels in VHL-WT compared to VHL-null RCC tumors. To examine the potential correlation between USP13 and pVHL status, we also depleted VHL expression by three independent single guide RNAs in pVHL-proficient HKC cells. VHL depletion led to up-regulation of USP13, corresponding with increased ZHX2 in these cells (SI Appendix, Fig. S6). Our findings suggest that VHL depletion may lead to increased USP13 that may contribute further to stabilizing ZHX2, which remains to be investigated for future research. Lastly, to further examine the relevance of USP13 and ZHX2 in ccRCC patients, we first examined their messenger RNA levels between normal and paired ccRCC tumors (n = 41) from the gene expression database available through University of Texas Southwestern Medical Center Kidney Cancer Specialized Program of Research Excellence (SPORE) (European Genome-Phenome Archive under the accession number EGAS00001005516) (35). Both ZHX2 and USP13 were highly expressed in tumors compared to normal controls (Fig. 6F). In addition, we also examined ZHX2 and USP13 mRNA levels from RNA sequencing data in patient-derived xenograft (PDX) available from the University of Texas Southwestern Medical Center Kidney SPORE program as described previously (36) and found that both ZHX2 and USP13 were highly expressed in PDX compared to paired normal tissue (Fig.6 G). In addition, USP13 and ZHX2 messenger RNA expression also correlated closely with one another, suggesting an independent regulatory mechanism other than ZHX2 protein stability controlled by USP13 in ccRCC patient samples (Fig. 6H). Taken together, our data strongly indicate that USP13 may play an important role in ccRCC pathogenesis.
Discussion
In our study, by performing a screen for potential ZHX2 DUBs, we identified USP13 as a potential DUB that regulates ZHX2 ubiquitination and protein stability. In addition, we showed that USP13 depletion led to decreased cell proliferation and 2D colony growth as well as 3D anchorage–independent growth in ccRCC. Depletion of USP13 led to decreased ccRCC tumorigenesis in vivo using an orthotopic xenograft model. Therefore, our findings suggest that USP13 may be a therapeutic target in ccRCC. Since USP13 regulates ZHX2 protein stability through its enzymatic activity, this study will motivate the development of USP13 inhibitors for potential treatment of ccRCC.
Here, we expand our understanding of the regulatory pathway of ZHX2. Previously, by using a genome-wide screening approach, we identified ZHX2 as a bona fide pVHL substrate (4). Its accumulation upon pVHL loss in ccRCC contributes to NF-κB activation and tumorigenesis (4). On the other hand, we also noted the heterogeneity in these ccRCC tumors even with VHL loss in our previous research (4). For example, some of ccRCC tumors with pVHL loss did not display up-regulation of ZHX2 (4), suggesting the additional layer of regulation on ZHX2 in a pVHL-independent manner. It is very likely in those tumors that ZHX2 may undergo additional layers of protein stability regulation by USP13 or other E3 ubiquitin ligases. Our current study identifies the DUB for ZHX2 in kidney cancer. On the other hand, it remains to be determined on what other pVHL-independent E3 ligases may regulate ZHX2 protein stability in kidney cancer, which is a potential caveat for our study (SI Appendix, Fig. S7).
The role of USP13 in cancer has been described previously. For example, USP13 was found to be amplified in lung cancer and ovarian cancer. The underlying molecular mechanism involves the bona fide USP13 substrates, including ATP citrate lyase, oxoglutarate dehydrogenase, and anti-apoptosis protein Mcl1 (37, 38). Specifically, one studied showed that USP13 depletion led to decreased MCL1 protein levels in lung cancer and ovarian cancer cells, therefore sensitizing tumor cells toward BH3 mimetic inhibitors (38). In another study, it was reported that USP13 mainly regulated ATP citrate lyase and oxoglutarate dehydrogenase, therefore affecting mitochondrial respiration, glutaminolysis, and fatty acid synthesis (37). It is likely that USP13 may regulate several substrates in lung and ovarian cancers, therefore providing a contributing role in those cancers. However, the role of USP13 in breast cancer appears to be as a tumor suppressor. Previous research showed that depletion of USP13 in breast cancer led to increased Akt phosphorylation, cell proliferation on 2D and 3D, glycolysis, and tumor growth by downregulating PTEN tumor suppressor proteins (25). In that study, USP13 was identified to be a DUB for PTEN and was found to be frequently lost in breast cancer (25). On the other hand, it remains unclear how USP13 was down-regulated in breast cancer. In addition, the role of USP13 as an oncogene or a tumor suppressor could be tumor type specific. Our clinical dataset in ccRCC showed that USP13 is overexpressed in tumor vs. normal controls. However, USP13 did not appear to be amplified in ccRCC. The previous research showed that the autophagy complex may interact with USP13 and regulate its protein stability (39). Given that autophagy was shown to be important for ccRCC tumor progression (40), it is possible that USP13 could be posttranscriptionally regulated in ccRCC, which remains to be further studied for future research. Since USP13 could regulate several substrates in other cancers including MCL1, SKP2, MITF, PTEN, TOPBP1, and c-MYC as described above, ZHX2 could be one of downstream targets for USP13 and other substrates could contribute to the lack of complete rescue in ccRCC, which remains to be further explored.
Spautin-1 was previously found to be the inhibitor for both USP10 and USP13 by promoting the degradation of Vps34 (39). Since USP10 is the DUB for p53, Spautin-1 also affects P53 in this setting (39). Interestingly, a recent unbiased screening identified Spautin-1 as a potent degrader for the IKZF1 transcription factor in an USP10- or USP13-independent fashion (41). Therefore, the role of Spautin-1 as an USP10/13 inhibitor needs further investigation. At this stage, due to the lack of USP13-specific inhibitors, we were unable to investigate the effect of pharmacological inhibition of USP13 in vivo. Nonetheless, our study hopefully will motivate the development of USP13-specific inhibitors that will be therapeutically beneficial in ccRCC.
Materials and Methods
Cell Culture and Reagents.
The 786-O, UMRC-2, UMRC-6, Caki-1, and 293T cells were purchased from the American Type Culture Collection (ATCC) or Sigma-Aldrich. OSRC2 cells were obtained from Cell Bank of Chinese Academy of Sciences. HKC cells were obtained from Dr. W. Kimryn Rathmell as described previously (32). The cell lines above were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco 11965118) with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (PS). All cells were cultured in an incubator at 37 °C with 5% CO2. All cells were verified by short tandem repeat testing. Mycoplasma detection was used to ensure cells were Mycoplasma free. MG132 was from Abcam (ab141003). Doxycycline was from Sigma-Aldrich (D9891). MTS assay (cell proliferation) was from Abcam (ab197010).
WB Analysis, Immunoprecipitation, and Antibodies.
EBC buffer (50 mM Tris base [pH 8.0], 0.5% Nonidet P-40, 0.1 mM EDTA, 120 mM sodium chloride (NaCl), and 10% glycerol) plus protease inhibitors and phosphatase inhibitors (Roche Applied bioscience) was used to lyse cells. The concentration of the cell lysate was measured via Protein Assay dye Reagent Concentrate (Bio-Rad, 500-0006). EBC buffer was used to adjust all cell lysates to the equal amount and concentration. Western blot analysis was conducted according to Sigma-Aldrich protocol (https://www.sigmaaldrich.com/JP/en/technical-documents/protocol/protein-biology/western-blotting/western-blotting). For immunoprecipitation, the lysates were mixed with HA beads (Roche Applied bioscience), FLAG beads (Roche Applied bioscience), or primary antibodies overnight. For primary antibodies, the lysate–antibody complex was incubated with protein G Sepharose beads (Roche Applied bioscience) for 4 h. The final lysate–protein–bead combinations were washed by EBC lysis 3 times and eluted by boiling in loading buffer. Bound proteins were resolved in polyacrylamide gel electrophoresis followed by western blot analysis.
Rabbit anti-USP13 (ab109264) and mouse anti-HIF2α (ab157249) were from Abcam. Mouse anti-Ubi (8017) and mouse anti-β-actin (sc-47778) were from Santa Cruz. Rabbit anti-ZHX2 (GTX112232) was from GeneTex. Rabbit anti-ZHX1 (A300-243A) was from Bethyl Laboratory. Rabbit anti-HA-tag (3724), rabbit anti-FLAG-tag (14793), rabbit-anti VHL (68547), mouse anti-His-tag (2366), and mouse anti-α-tubulin (3873S) were from Cell Signaling Technology. Mouse anti-GAPDH (TA-08) was from Zhongshan Gold Bridge Bio. Anti-FLAG M2 Affinity Gel (A2220) and Anti-HA Affinity Matrix (11815016001) were from Sigma-Aldrich. Antibodies used for IHC stating were mouse anti-USP13 (B-9) (Santa Cruz, sc-514416) and Rabbit anti-ZHX2 (GeneTex, GTX112232).
Plasmids, Virus Production, and Infection.
DUB cDNA plasmids and the library were from Addgene. The USP13 catalytic dead mutant (C345A) was kindly provided by Dr. Shideng Bao’s laboratory from Cleveland Clinic as described previously (28). The USP13 complete coding sequence and different ZHX2 truncation mutants’ sequences were cloned into a pLenti-6 FLAG lentivirus vector according to the standard procedure. USP13 short hairpin RNA-resistance plasmids were generated by site-directed mutagenesis (Agilent Technologies). Short hairpin RNA resistance sequences are as follows: Sh50 resistant, GATGACTCAAAAGATCGGCTCATGAATCAGTTAATTGACCCATCAGACATCG; and Sh52 resistant, CAGTATTCAAAGCCTCCCGTCAAGTCAGAGCTGATAGAACAGGTGATGAAG. Virus production and infection were conducted according to the standard protocol.
siRNAs, Lentiviral shRNA, and sgRNA Vectors.
USP13 short hairpin RNAs (42, 43) were purchased from Sigma Aldrich. USP13 siRNAs were purchased from Dharmacon (D006064). Single guide RNAs targeting USP13 were inserted into the CRISPR-V2 lentivirus vector. Short hairpin RNAs targeting USP13 were cloned into the tet-on puro vector to generate doxycycline-inducible shUSP13 plasmids. shRNA and single guide RNA sequences are as follows: Sg1, AGCGCCGGGGCGCCCTGTT; Sg2, AATAGCACTACCAAATATTG; Sg3, TGTATGCATGAATACATTTT; Sg4, TGCCCACGATCCGCGTGCCC; Sh50, CGCCTGATGAACCAATTGATA; and Sh52, CCGGTGAAATCTGAACTCATT.
Ubiquitination Assay.
Next, 1% SDS lysis buffer was used to lyse cells followed by boiling in a heater for 10 min. Ten times the volume of EBC buffer (complete protease inhibitors, phosphatase inhibitors, and 20 mM N-ethylmaleimide ([E3876, Sigma]) was used to dilute cell lysates. Then diluted lysates was sonicated and incubated with primary antibodies or NTA-beads overnight. The subsequent steps are same as immunoblotting analysis above.
RNA-Seq.
Total RNA from 786-O cells (triplicates) was extracted by using an RNeasy kit with on-column DNase digestion (Qiagen). Libraries were prepared using the TruSeq RNA Library Prep Kit v2 (Illumina) according to the manufacturer’s instructions. Samples were sequenced at Novogene on a NovaSeq 6000 with paired-end 150-bp reads. Reads were then filtered for adapter contamination using cutadapt (44) and filtered using the FASTX-Toolkit (v0.0.14) (hannonlab.cshl.edu/fastx_toolkit/index.html) such that at least 90% of bases of each read had a Phred score of >20. Reads were aligned to the reference genome (hg19) using STAR (v2.5.2b) by retaining only primary alignments (42). Reads overlapping blacklisted regions of the genome were then removed. Transcript abundance was then estimated using salmon (v0.11.3), and differential expression was detected using DESeq2 (v1.14.1) (45). To call differentially expressed genes following either USP13 or ZHX2 silencing, both short hairpin RNAs for a given target were combined into one group and compared against the paired shCtrl samples using DESeq2. Python (v3.7.9) was used for data filtering and formatting as well as making volcano plots. Heatmaps were plotted with Morpheus (https://software.broadinstitute.org/morpheus). Heatmaps were scaled by subtracting each value in a row by the per-row mean of the shCtrl replicates. Variance-stabilizing transformed expression values were used for plotting. Pathway enrichment was performed using gProfiler (43).
Real-Time PCR.
Extraction of total RNA from cells using TRIzol reagent (Invitrogen) was performed according to the manufacturer’s instruction. Reverse transcription total RNA was performed using an iScript cDNA synthesis kit (Bio-Rad). RT-qPCR was performed using a QuantiTect SYBR Green PCR master mix (Qiagen) according to the manufacturer’s instructions. Relative expression levels were calculated using the 2(-delta delta threshold cycle) (2-ΔΔCt) method. The transcription level of GAPDH serves as an internal reference.
Cell Proliferation Assays.
Cells were planted in 5 replicates in 96-well plates (500 cells/well) in 200 µL growth medium. At the indicated time points, cells were changed with 180 µL fresh growth medium supplemented with 20 µL MTS reagents at 37 °C for 1 h. The optical density value was detected at 490 nm using a 96-well plate reader (Tecan).
2D Colony Formation Assay and Anchorage-Independent Growth Assays.
For the 2D colony formation assay, 2,000 cells were planted in a 6-well plate after infection or transfection. Medium was changed every 3 d. When one of the wells grew full, cells were fixed by 5% methanol for 20 min and stained with crystal violet for 20 min. A stained colony was washed by phosphate-buffered saline (PBS) followed by imaging. For the 3D anchorage-independent growth assay, 5,000 or 10,000 cells were planted on DMEM with 0.4% agarose (Fisher Scientific, BP165-25) onto bottom layers (DMEM with 1% agarose). The medium above the top layers was changed every 3 or 4 d. When the 3D colony grew to a sufficient size, colonies were stained with iodonitrotetrazolium chloride solution.
Tumor Xenograft and Orthotopic Tumor Growth.
A total of 5 × 106 stable OSRC-2 cells were planted into the axilla of 5-wk-old BALB/c nude mice (Vital River Laboratory Animal Technology Co., Ltd. No.401). When the length of a tumor reached over 2 mm, the tumor size was recorded per week. Tumor samples were collected when the length of the tumor reach 2 cm. After the tumor tissue was ground, the protein was extracted for western blot.
Next, 5-wk-old NSG (Jackson laboratory, male: female = 2:1) mice were used for orthotopic tumor xenograft studies. Approximately 5 × 105 OSRC-2 or 786-O cells expressing tet-on shCtrl or shUSP13 were resuspended in 20 μL DMEM with 2% FBS and planted into the kidney capsule of the NSG mouse kidney. Bioluminescence imaging was performed as described previously (46). After 1 or 2 wk, mice were fed with rodent chow with doxycycline (Beyotime, ST039A). Mice were euthanized after 7 wk, and lungs were removed followed by imaging to examine tumor metastasis. The weight of tumors and bioluminescence intensity were presented as mean ± SEM and evaluated statistically using the t test. OSRC-2 animal experiments were approved by Peking University First Hospital Institutional Animal Care and Use Committee (IACUC). All other animal experiments followed NIH guidelines and were approved by the University of Texas Southwestern Medical Center IACUC.
Human ccRCC Tumors and Immunohistochemistry Staining.
Fresh–frozen samples or tissue microarrays (TMAs) of ccRCC and kidney-adjacent normal tissue were obtained from The Tissue Management Shared Resource in University of Texas Southwestern Medical Center or Dr. Gong’s laboratory at Peking University. TMA contains 153 pairs of tumor and normal tissue of ccRCC patients from Peking University First Hospital, and all the pathological results were diagnosed by the Department of Pathology, Peking University First Hospital, Institute of Urology. All human tumor sample–related studies have been approved by the University of Texas Southwestern Medical Center or University of North Carolina Chapel Hill Institutional Review Boards, which were all deidentified human patient samples. TMAs were immunostained as described previously (4). Briefly, slides were dewaxed and hydrated. Heat-induced antigen retrieval was performed for TMAs followed by blocking. After pretreatment, TMAs were incubated with rabbit polyclonal anti-ZHX2 antibody (1:100, GeneTex, GTX112232) or mouse monoclonal anti-USP13 antibody (1:20, Santa Cruz, sc514416) overnight, and then TMAs were stained by DAB. 3D histech (MRXS) was used to scan the stained TMA slides. Then we used Quant Center, which is provided with the 3D histech scanner, to score the number of positives and only the brown-yellow color stained by DAB can be quantified. H-SCORE is the abbreviation of histochemistry score. The higher the H-SCORE value, the higher the staining intensity. It is a histological scoring method for processing the results of immunohistochemistry. The number of positive cells in each section and the staining intensity are converted into corresponding values to achieve the purpose of staining semiquantitatively. H-SCORE = ∑(PI × I) = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + percentage of cells of strong intensity × 3), where PI represents the number of positive cells and I represents the intensity of staining.
Cell Apoptosis Assay and Cell Cycle Analysis.
Cell apoptosis was assayed by staining with the FITC Annexin V Apoptosis Detection Kit with PI (BiolegendSupplier Diversity Partner, No. 50-403-826) following the manufacturer’s instructions. For the cell cycle assays, cells were harvested in the appropriate manner and washed in phosphate-buffered saline. The cells were fixed in cold 70% ethanol for 30 min at 4 °C, and then were washed twice with phosphate-buffered saline. The cells were spun at 850 g in a centrifuge. Supernatants were discarded, and the cells were treated with ribonuclease, 50 μL of a 100-μg/mL stock of RNase, and 200 μL PI (from 50 -μg/mL stock solution) for 15 min at room temperature. Samples were then analyzed for their DNA content using flow cytometry.
OCR Measurement.
The OCR experiment was described in detail previously with some modifications (34). Briefly, cells were seeded into the XF24 cell culture microplate in a density of ∼3 × 104 cells/well, with 5∼7 technical replicates for each cell line. The cells were cultured in the substrate-limited medium (basal DMEM [Life Technologies, #A14430-01] supplemented with 0.5 mM glucose, 1.0 mM GlutaMAX [Life Technologies, #35050061], 0.5 mM Carnitine, and 1% FBS) overnight before the assay. The next day, the medium was changed with the fatty acid oxidation (FAO) assay buffer (111 mM sodium chloride, 4.7 mM KCl, 1.25 mM CaCl2, 2.0 mM MgSO4, 1.2 mM NaH2PO4) supplemented with 2.5 mM glucose, 0.5 mM carnitine, and 5 mM Hepes, and pH was adjusted to 7.4. The Palmitate-BSA substrate (Agilent, 102720-100) was added to each well before the assay running. OCR were measured by an XFe24 extracellular flux analyzer (Agilent Technologies), according to the manufacturer’s instructions. Data were normalized to the actual cell number counted after the machine measurement.
Statistical Analysis.
Unless indicated, SPSS or GraphPad Prism were used to analyze the statistical significances of data. All graphs depict mean ± SEM unless otherwise indicated. *, ** and *** represent P value of <0.05, 0.01, and 0.001, respectively. n.s. represents not significant. GraphPad Prism was used to generate all the graphs.
Supplementary Material
Acknowledgments
This work was supported in part by the National Cancer Institute (NCI) (Q.Z., R01CA211732, R01CA256833, and R21CA223675) and Cancer Prevention and Research Institute of Texas (CPRIT, RR190058 to Q.Z). Q.Z. was also supported by Developmental Research Program from Kidney Cancer SPORE at University of Texas Southwestern Medical Center sponsored by NCI (P50CA196516). J.M.S. and A.J.H. were supported by The Eunice Kennedy Shriver National Institute of Child Health and Human Development (U54HD079124) and National Institute of Neurological Disorders and Stroke (P30NS045892). Q.Z is an American Cancer Society Research Scholar, CPRIT Scholar in Cancer Research, V Scholar, Kimmel Scholar, Susan G. Komen Career Catalyst awardee, and Mary Kay Foundation awardee. Q.Z is also supported by Kidney Cancer Research Alliance (KCCure).
Footnotes
Competing interest statement: Q.Z. received consultation fee from Bristol Myers Squibb (BMS), which is unrelated to this study. Other authors do not have any conflicts.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119854119/-/DCSupplemental.
Data, Materials, and Software Availability
RNA sequencing data have been deposited to the Gene Expression Omnibus under accession number GSE186717 (47). The data with accession no. GSE186717 are public available.
References
- 1.Kondo K., Klco J., Nakamura E., Lechpammer M., Kaelin W. G. Jr., Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell 1, 237–246 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Ivan M., et al. , HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 292, 464–468 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P., et al. , Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., et al. , VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 361, 290–295 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., et al. , Genome-wide screening identifies SFMBT1 as an oncogenic driver in cancer with VHL loss. Mol. Cell 77, 1294–1306.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Kaelin W. G. Jr., Molecular pathogenesis of the von Hippel-Lindau hereditary cancer syndrome: Implications for oxygen sensing. Cell Growth Differ. 12, 447–455 (2001). [PubMed] [Google Scholar]
- 7.Raval R. R., et al. , Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25, 5675–5686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo K., Kim W. Y., Lechpammer M., Kaelin W. G. Jr., Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong K., et al. , USP37 promotes deubiquitination of HIF2α in kidney cancer. Proc. Natl. Acad. Sci. U.S.A. 117, 13023–13032 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho H., et al. , On-target efficacy of a HIF2alpha antagonist in preclinical kidney cancer models. Nature (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., et al. , Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada K., Printz R. L., Osawa H., Granner D. K., Human ZHX1: Cloning, chromosomal location, and interaction with transcription factor NF-Y. Biochem. Biophys. Res. Commun. 261, 614–621 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Kawata H., et al. , Zinc-fingers and homeoboxes (ZHX) 2, a novel member of the ZHX family, functions as a transcriptional repressor. Biochem. J. 373, 747–757 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yue X., et al. , Zinc fingers and homeoboxes 2 inhibits hepatocellular carcinoma cell proliferation and represses expression of Cyclins A and E. Gastroenterology 142, 1559–1570 e1552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma H., et al. , ZHX2 enhances the cytotoxicity of chemotherapeutic drugs in liver tumor cells by repressing MDR1 via interfering with NF-YA. Oncotarget 6, 1049–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S., et al. , Expression of zinc-fingers and homeoboxes 2 in hepatocellular carcinogenesis: A tissue microarray and clinicopathological analysis. Neoplasma 54, 207–211 (2007). [PubMed] [Google Scholar]
- 17.Cheng A., Guo X., Dai X., Wang Z., Upregulation of ZHX2 predicts poor prognosis and is correlated with immune infiltration in gastric cancer. FEBS Open Bio 11, 1785–1798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., et al. , Novel VHL substrate targets SFMBT1 and ZHX2 may be important prognostic predictors in patients with ccRCC. Oncol. Lett. 21, 379 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang J., et al. , ZHX2 mediates proteasome inhibitor resistance via regulating nuclear translocation of NF-κB in multiple myeloma. Cancer Med. 9, 7244–7252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu L., Ding R., Yan H., Zhang J., Lin Z., ZHX2 drives cell growth and migration via activating MEK/ERK signal and induces Sunitinib resistance by regulating the autophagy in clear cell renal cell carcinoma. Cell Death Dis. 11, 337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang W., et al. , ZHX2 promotes HIF1α oncogenic signaling in triple-negative breast cancer. eLife 10, 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan Y., et al. , Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin. Cancer Res. 13, 5745–5755 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W., Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan E. L., et al. , The deubiquitinase (DUB) USP13 promotes Mcl-1 stabilisation in cervical cancer. Oncogene 40, 2112–2129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., et al. , Deubiquitylation and stabilization of PTEN by USP13. Nat. Cell Biol. 15, 1486–1494 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M., Gutierrez G. J., Ronai Z. A., Ubiquitin-recognition protein Ufd1 couples the endoplasmic reticulum (ER) stress response to cell cycle control. Proc. Natl. Acad. Sci. U.S.A. 108, 9119–9124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X., Fiske B., Kawakami A., Li J., Fisher D. E., Regulation of MITF stability by the USP13 deubiquitinase. Nat. Commun. 2, 414 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Fang X., et al. , Deubiquitinase USP13 maintains glioblastoma stem cells by antagonizing FBXL14-mediated Myc ubiquitination. J. Exp. Med. 214, 245–267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim W., et al. , USP13 regulates the replication stress response by deubiquitinating TopBP1. DNA Repair (Amst.) 100, 103063 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scortegagna M., et al. , USP13 enzyme regulates Siah2 ligase stability and activity via noncatalytic ubiquitin-binding domains. J. Biol. Chem. 286, 27333–27341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu B., et al. , HIF2α-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear-cell renal cell carcinoma. Cancer Discov. 5, 652–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu L., et al. , TBK1 is a synthetic lethal target in cancer with VHL loss. Cancer Discov. 10, 460–475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Detre S., Saclani Jotti G., Dowsett M., A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 48, 876–878 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao C., et al. , Identification of BBOX1 as a therapeutic target in triple-negative breast cancer. Cancer Discov. 10, 1706–1721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elias R., et al. , A renal cell carcinoma tumorgraft platform to advance precision medicine. Cell Rep. 37, 110055 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivanand S., et al. , A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci. Transl. Med. 4, 137ra75 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han C., et al. , Amplification of USP13 drives ovarian cancer metabolism. Nat. Commun. 7, 13525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S., et al. , Deubiquitinase USP13 dictates MCL1 stability and sensitivity to BH3 mimetic inhibitors. Nat. Commun. 9, 215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J., et al. , Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikhaylova O., et al. , VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell 21, 532–546 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koduri V., et al. , Targeting oncoproteins with a positive selection assay for protein degraders. Sci. Adv. 7, eabd6263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reimand J., et al. , Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 14, 482–517 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3 (2011). [Google Scholar]
- 45.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q., et al. , Control of cyclin D1 and breast tumorigenesis by the EglN2 prolyl hydroxylase. Cancer Cell 16, 413–424 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie H., et al., USP13 promotes deubiquitination of ZHX2 and tumorigenesis in kidney cancer. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE186717. Deposited 27 October 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited to the Gene Expression Omnibus under accession number GSE186717 (47). The data with accession no. GSE186717 are public available.