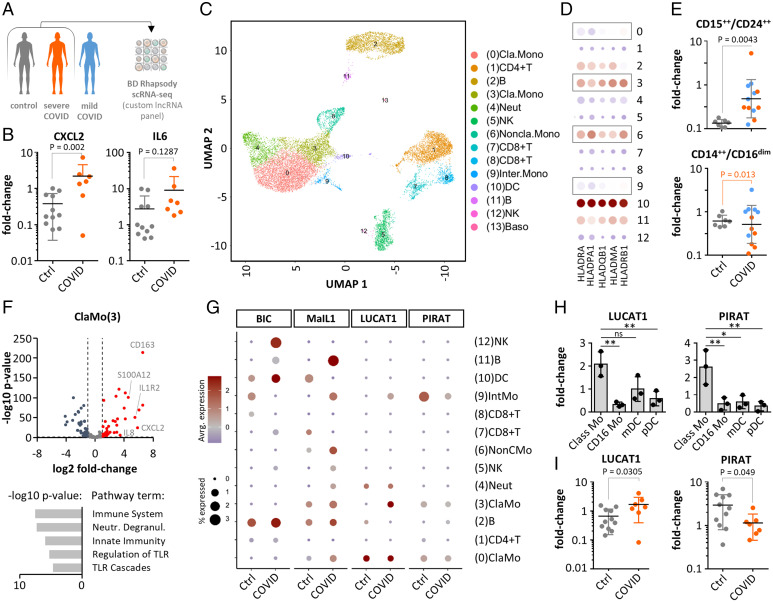
Fig. 2.
scRNA-seq analysis of lincRNA expression during COVID-19. (A) Patient PBMC analysis strategy. (B) Validation of immune marker induction in COVID-19 cohort PBMCs (qRT-PCR, control-patient 1 set as reference). (C) UMAP-plot with color-coded cell populations identified in merged scRNA-seq data. (D) HLA mRNA expression profile (scRNA-seq, monocytes highlighted). (E) FACS validation of immature neutrophil (CD15++/CD24++) appearance and reduction of classic monocytes (CD14++/CD16dim) in COVID-19 (color-coded according to A). (F) Volcano plot (Upper) and Reactome pathway (Lower) analysis of classic monocyte response during COVID-19 (scRNA-seq). (G) LincRNA profiles in control and COVID-19 patients (scRNA-seq). (H) PIRAT and LUCAT1 expression in classic (Class) and nonclassic (CD16) monocytes, myeloid dendritic cells (mDC), and plasmacytoid DCs (pDC) (qRT-PCR). (I) Same as B, but for PIRAT and LUCAT1. (B, E, and I) Two-tailed Student’s t test. (H) One-way ANOVA, three independent experiments. *P ≤ 0.05; **P ≤ 0.01.