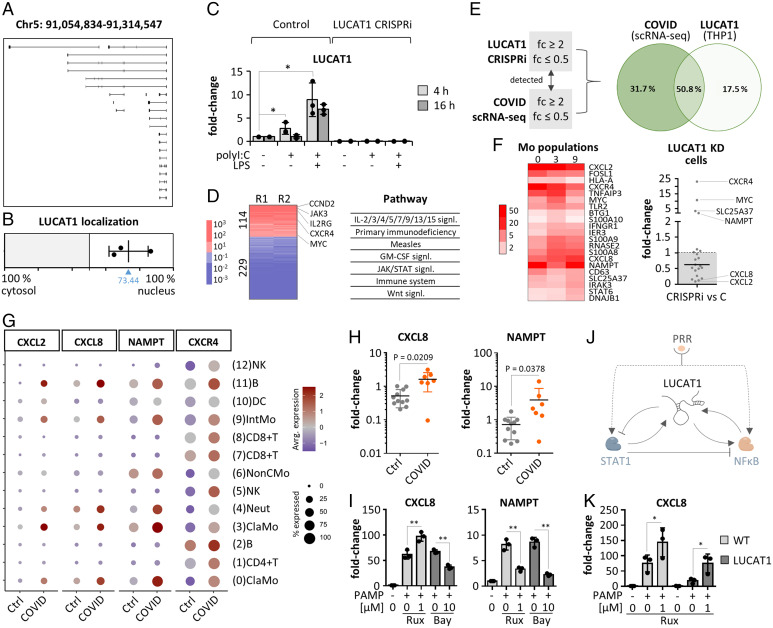
Fig. 3.
Role of LUCAT1 in monocytes. (A) ENSEMBL-annotated LUCAT1 isoforms. (B) LUCAT1 subcellular localization (qRT-PCR). (C) LUCAT1 expression in control and knockdown THP1 monocytes (qRT-PCR, relative to unstimulated control). (D) RNA-seq analysis (transcripts regulated ≥10-fold, Kyoto Encyclopedia of Genes and Genomes/Reactome pathways) of LUCAT1 knockdown versus control THP1 monocytes, activated for 4 h with polyI:C and LPS. R1 and R2 = replicates 1 and 2. (E) Overlap of gene regulations twofold or greater (up or down) in monocytes during COVID-19 (scRNA-seq populations 0, 3, and 9) (Fig. 2C) and upon LUCAT1 knockdown in THP1 cells. (F, Left) COVID-19-induced (twofold or greater) mRNAs in monocyte populations from Fig. 2C. (Right) Regulation of the same mRNAs in dataset from D. (G) Expression of LUCAT1-controlled mRNAs in scRNA-seq data. (H) Same as Fig. 2B, but for CXCL8 and NAMPT. (I) Ruxolitinib and BAY-11-7082 sensitivity of selected mRNAs (monocytes; PAMP = 4 h LPS + polyI:C). Fold-changes relative to unstimulated control. (J) Model of LUCAT1 function. (K) Rescue of CXCL8 dysregulation in LUCAT1-deficient THP1 cells upon 2-h Ruxolitinib pretreatment. (C, I, and K) One-way ANOVA, three independent experiments. (H) Two-tailed Student’s t test. *P ≤ 0.05; **P ≤ 0.01.