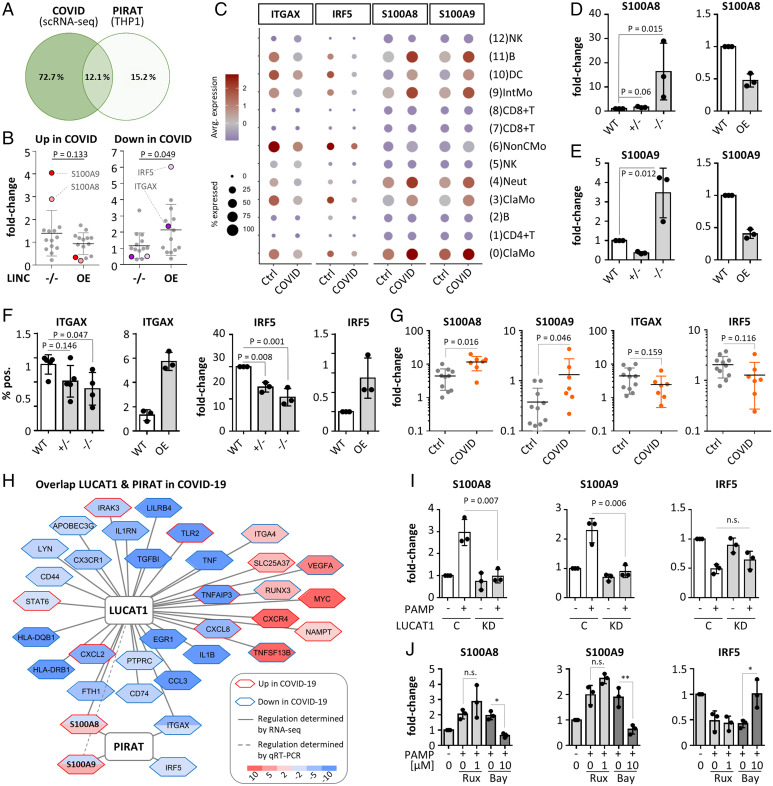
Fig. 5.
Participation of PIRAT in PU.1 circuits relevant to COVID-19. (A) Overlap of genes regulated twofold or greater in classic monocytes during COVID-19 (scRNA-seq data) and upon PIRAT knockout/overexpression (data from Fig. 4G). (B) Regulation of genes in PIRAT knockout (−/−) or overexpression (OE) compared to wild-type cells, up-regulated (Left) or down-regulated (Right) during COVID-19. (C) Cell-type–specific expression of top PIRAT-controlled, COVID-responsive mRNAs from B (scRNA-seq). (D–F) Expression changes of PIRAT-controlled PU.1 targets in PIRAT knockout and overexpression compared to wild-type THP1 cells (qRT-PCR). (G) Regulation of PIRAT-controlled genes in PBMCs from COVID-19 and control-patients (qRT-PCR, control-patient 1 set as reference). (H) Overlap of LUCAT1 and PIRAT controlled genes (THP1 RNA-seq data from A and Fig. 3E), regulated twofold or greater (up or down). Fill-colors indicate regulations due to lncRNA deficiency. Plot reduced to genes regulated twofold or greater (up or down) during COVID-19 (scRNA-seq, Fig. 2). (I) Regulation of S100A8, S100A9, and IRF5 in control and LUCAT1-CRISPRi cells from Fig. 3 (4 h LPS and polyI:C). (J) Same as Fig. 3I, but for S100A8, S100A9, and IRF5. (D–F and I and J) One-way ANOVA; (G) two-tailed Student’s t test; ≥3 independent experiments. *P ≤ 0.05; **P ≤ 0.01.