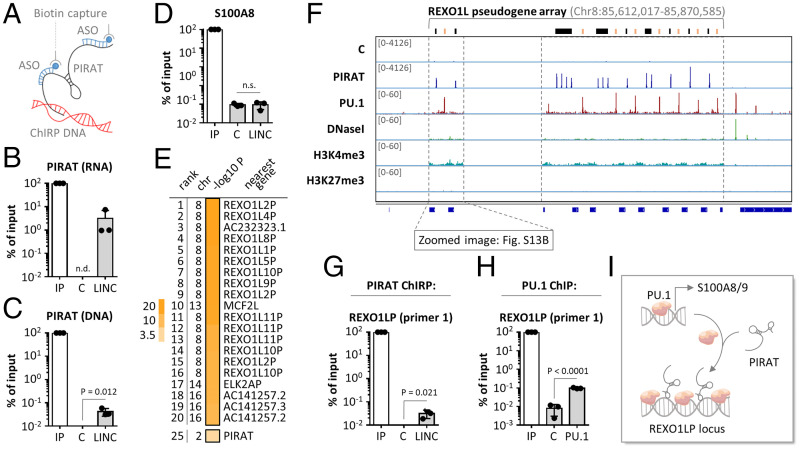
Fig. 6.
Repetitive binding of PU.1 and PIRAT to the REXO1LP locus. (A) PIRAT ChIRP was performed using primary CD14+ monocytes. (B) Recovery of PIRAT RNA in ChIRP samples compared to input (IP) sample (C = control ChIRP, LINC = PIRAT ChIRP; qRT-PCR). (C) Same as B but with genomic DNA. (D) Same as C but with S100A8 promoter detection. (E) Summary of PIRAT binding site peak-calling (ChIRP-seq; chr = chromosome; top 20 peaks and peak #25 are shown; full list of peaks in SI Appendix, Table S5). (F) IGV plots showing control (C) and PIRAT ChIRP-seq and matched CD14+-monocyte PU.1 ChIP-, DNaseI-, and histone-3 ChIP-seq coverage in the REXO1LP locus. Track-heightin brackets. (G) qRT-PCR validation of PIRAT binding to ChIRP peaks in the REXO1LP locus. (H) qRT-PCR validation of PU.1 binding to ChIP peaks in the REXO1LP locus. (I) Model of PU.1 redirection from alarmin promoters to the REXO1LP locus by PIRAT. (B–D, G, and H) Two-tailed Student’s t test, three independent experiments.