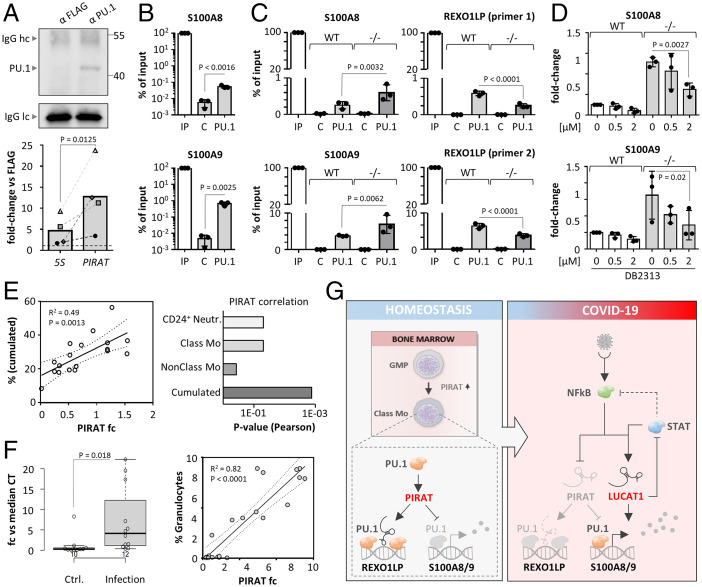
Fig. 7.
PIRAT redirects PU.1 from alarmin promoters to the REXO1LP repeat. (A) Western blot validation of PU.1 capture (Upper; hc/lc = light chain/heavy chain) and qRT-PCR analysis of PIRAT enrichment (Lower) in PU.1 UV-CLIP (primary monocytes). (B) ChIP qRT-PCR analysis of PU.1 binding to S100A8 and A9 promoter DNA in primary monocytes. (C) qRT-PCR analysis of PU.1 binding to ChIP-seq peaks in the S100A8 and S1009 promoters and the REXO1LP locus, in wild-type (WT) and PIRAT knockout (−/−) THP1 cells. (D) qRT-PCR analysis of S100A8 and S100A9 expression after treatment of wild-type (WT) and PIRAT knockout (−/−) THP1 cells with PU.1 inhibitor DB2313 (concentrations indicated) for 4 h. (E, Left) Pearson correlation of cumulated neutrophil and classic monocyte percentage with PIRAT levels in COVID-19 patient PBMCs. (Right) P values for PIRAT correlation (Pearson) with percentages of the indicated cell types in COVID-19 patient PBMCs. (F, Left) qRT-PCR analysis of PIRAT expression in BALF from control and pulmonary infection patients. (Right) Pearson correlation of granulocyte percentage with PIRAT expression in BALF. (G) Summary of alarmin-control by PIRAT and LUCAT1 in monocytes. (A, B, and F) Two-tailed Student’s t test. (C and D) One-way ANOVA; ≥3 independent experiments were performed. Abbreviations: C, control; IP, input.