Figure 3.
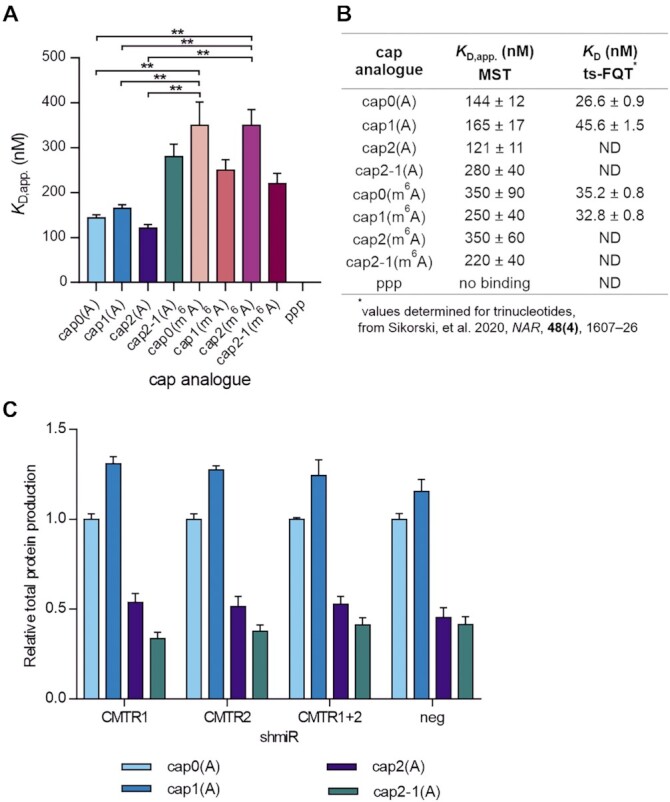
2′-O-Methylation within the cap structure does not influence RNA affinity for the translational machinery. (A) Relative affinities of transcripts bearing different cap analogues for murine eIF4E determined using MST. Bars represent the mean value ± SEM from three independent replicates. Statistical significance: **P <0.01 (one-way ANOVA with Turkey's multiple comparisons test). (Representative MST curves and competitive binding curves obtained in the experiment are presented in Supplementary Figure S8.) (B) Comparison of apparent binding constant values KD,app for capped RNAs in complexes with murine eIF4E measured with MST with dissociation constants of eIF4E–cap complexes obtained using time-synchronized fluorescence quenching titration (ts-FQT) (14). For all RNAs, three independent replicates were performed, besides cap2(A) and cap2-1(A) RNAs, for which data from two replicates are presented. (C) Competition of differently capped IVT mRNAs encoding Gaussia luciferase with endogenous mRNAs for the translation machinery. HEK 293 Flp-In T-REx cells expressing shmiRs targeting CMTR1, CMTR2 or both (a negative control was utilized in parallel) were transfected with IVT mRNAs, and medium was collected after 72 h for luciferase activity analysis. Bars represent the mean value ± SEM normalized to Gaussia luciferase activity measured for transcripts with cap0(A). Data for three independent experiments are presented (each biological replicate consisted of three transfections).
