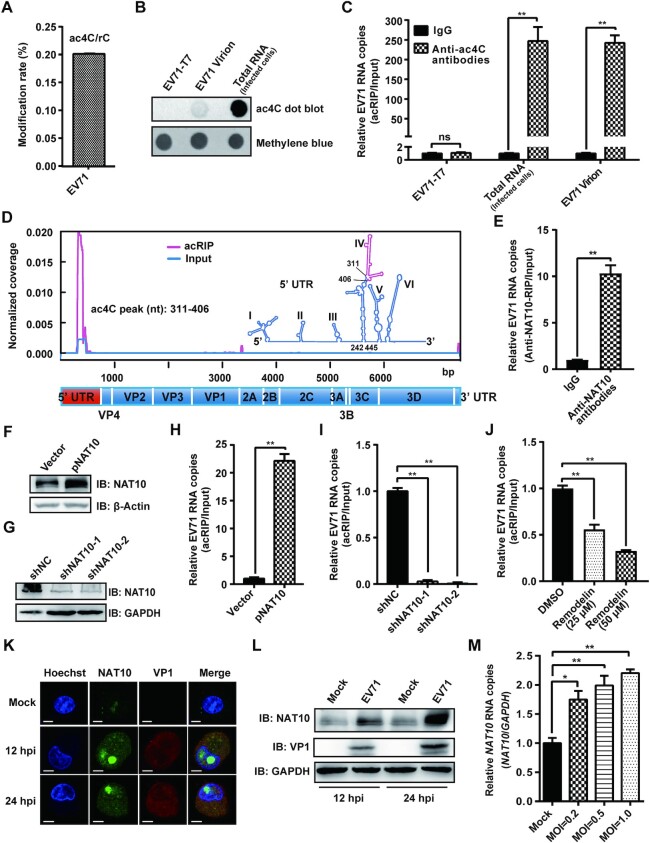
Figure 1.
EV71 contained the ac4C modification and altered the expression pattern of NAT10. (A) UPLC-MS/MS of the EV71 RNA genome. EV71 RNA was extracted from viral supernatants concentrated by ultracentrifugation. The percentage of ac4C among C residues is presented on the y-axis. (B) Anti-ac4C dot blot of EV71 RNA from (A). Methylene blue staining was used as a loading control. The in vitro T7-transcribed EV71 genome and total RNA from EV71-infected Vero cells served as negative and positive controls, respectively. (C) acRIP-qRT-PCR. RNAs from EV71-infected cells, EV71 virions (A), and transcribed by T7 polymerase were incubated with IgG or anti-ac4C antibodies, followed by IP and qRT-PCR. Data are means ± SEMs (n = 3). **P ≤ 0.01, ns: not significant, unpaired Student's t-tests. (D) Mapping of ac4C sites on EV71 RNA by acRIP-Seq. Poly(A)+ RNA, purified from total RNA extracted from EV71-infected Vero cells, was fragmented, and IP was performed using anti-ac4C antibodies, followed by next-generation sequencing. Normalized coverage on acRIP-Seq and input EV71 RNA are presented in red and blue, respectively. The RNA secondary structure was predicted using Mfold software. The red line in stem–loop IV represents the peak area of ac4C. (E) Binding of NAT10 to EV71 RNA. EV71-infected Vero cells were crosslinked using formaldehyde, and IP was performed using anti-NAT10 antibodies. The results were quantified by qPCR. IgG was used as a negative control. Data are means ± SEMs (n = 3). **P ≤ 0.01, unpaired Student's t-tests. (F, G) Western blotting of extracts from EV71-infected Vero cells treated with pNAT10 (F) or shRNA (G). The expression of NAT10 was assessed using anti-NAT10 antibodies, and GAPDH served as a loading control. (H–J) Modulation of EV71 RNA acetylation by NAT10. Total RNA was extracted from EV71-infected Vero cells with NAT10 overexpression (H), knockdown (I), or inhibition (J); isolated using acRIP; and quantified using qRT-PCR. Data are means ± SEMs (n = 3). **P ≤ 0.01, unpaired Student's t-tests. (K) Confocal microscopy images of NAT10 in EV71- or mock-infected cells. The nucleus (blue), EV71 protein (red), and NAT10 (green) were labeled with Hoechst 33258, anti-VP1 antibodies, and anti-NAT10 antibodies, respectively. Scale bars, 5 μm. (L) Western blot analysis of extracts of EV71-infected (MOI = 1) Vero cells (12 and 24 hpi). GAPDH was used as a loading control. (M) RNA expression levels of NAT10. Total RNA was extracted at 12 hpi from EV71- (MOI = 0.2, 0.5, or 1) or mock-infected Vero cells and quantified using qRT-PCR. GAPDH was used as a control. Data are means ± SEMs (n = 3). *P ≤ 0.05, **P ≤ 0.01, unpaired Student's t-tests.