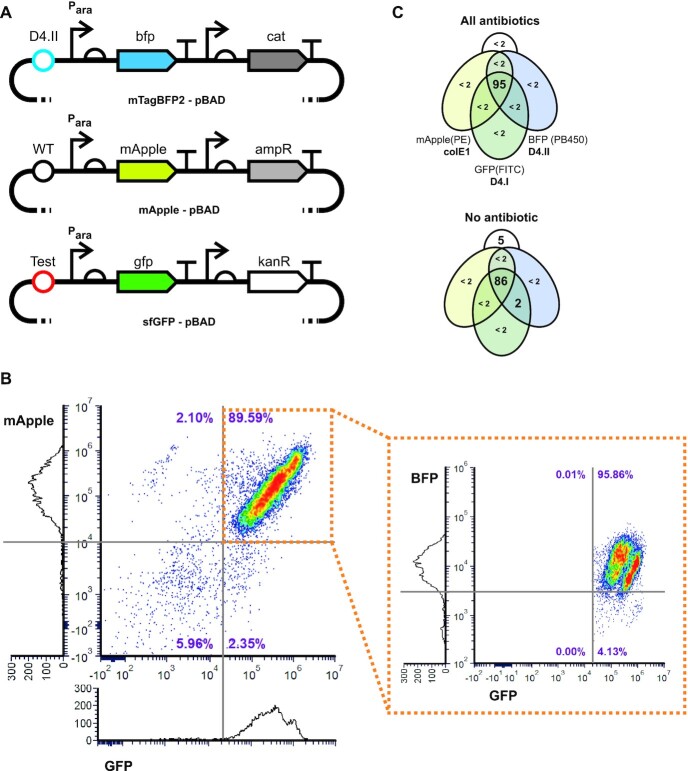
Figure 5.
Selected plasmid origins intercompatibility. (A) pBAD vectors harbouring fluorescent protein genes (based on plasmids # 54519, #54536 and #54572 from Addgene) were engineered to harbour different replication origins and different selectable markers. (B) Flow cytometry analysis of bacteria harbouring colE1, D4.I and D4.II origins after overnight growth in the absence of any antibiotic selection. Cells expressing both GFP and mApple are gated (orange box) and checked for BFP expression (inset dot plot). Histograms of individual channels (without gating) are shown. (C) Flow cytometry results are summarized in the Venn diagrams, showing all subsets above 2% of the population to the nearest percentage point. Rare combinations (below 2% of the total population) are shown as ‘<2’ for clarity. Detection events without fluorescence (e.g. dead cells or residual electronic noise) are shown in the peripheral (white) set. For the colE1, D4.I and D4.II combination, even in the absence of antibiotics, all plasmids remained stably in the cells in contrast to the two-plasmid cross-compatibility assays.