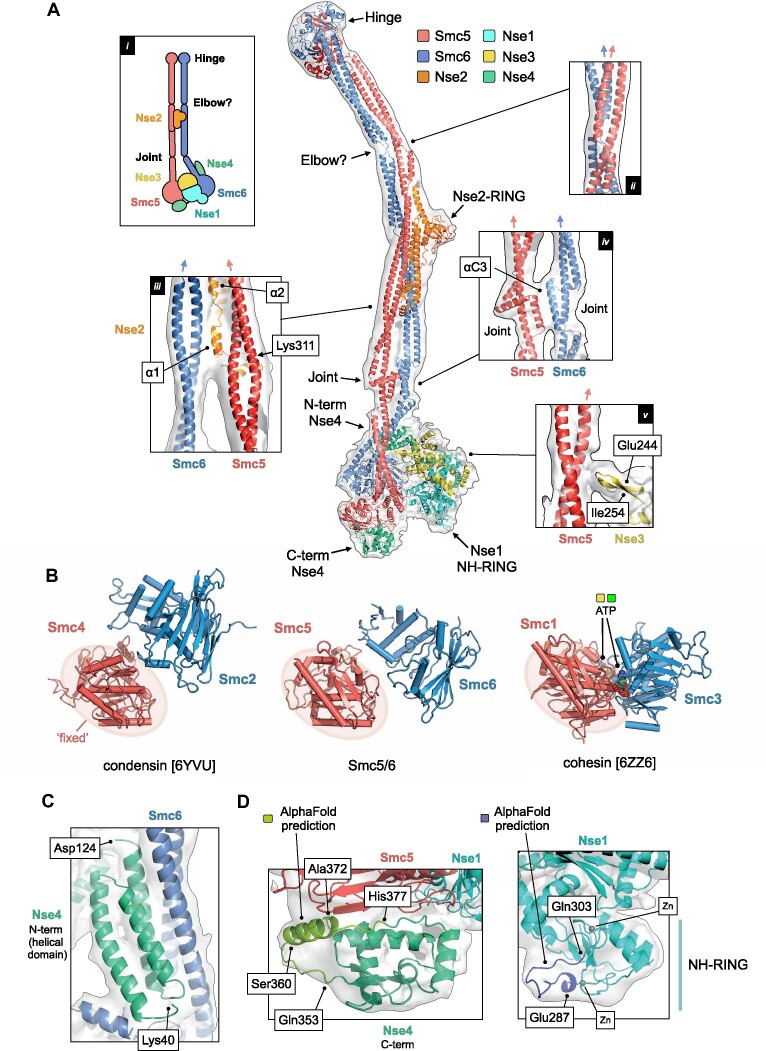
Figure 2.
A pseudo-atomic model for the Smc5/6 holo-complex. (A) Overview of the pseudo-atomic model. (A, inset i) Schematic showing the overall architecture of the Smc5/6 complex and selected molecular features. (A, inset ii) Expanded view of the ‘Elbow’, highlighting the crossover of the coiled-coil ‘arms’ of Smc5 and Smc6 at this point. (A, inset iii) The first alpha-helix of Nse2 (α1) is situated between the two arms of the complex. The position of Lys311, a known site of auto-SUMOylation is also indicated (A, inset iv) Expanded view of the interface between the ‘Joint’ features of Smc5 and Smc6, involving the two αC3 helices. (A, inset v) A short beta-hairpin (amino acids Glu244-Ile254) protruding from Nse3 is in close proximity to the arm of Smc5. For each inset, the directionality of the ascending helix (head to hinge) is indicated by a blue or red arrow, for Smc5 and Smc6 respectively. (B) Comparison of the relative head domain positions in the cryo-EM structures of budding yeast condensin (PDB: 6YVU), Smc5/6 (this manuscript) and cohesin (PDB: 6ZZ6); in each, using the head of the κ-SMC as a fixed reference point. (C) Expanded view for the N-terminal helical domain of Nse4 (aa Lys40-Asp124) bound to the ‘arm’ of Smc6. (D, left) AlphaFold predicts the presence of an additional helical element (aa Ser360-Ala372) in the C-terminal domain of Nse4. (D, right). AlphaFold predicts a budding yeast-specific loop insertion in the NH-RING of Nse1 (aa Glu287-Gln303). Where shown, sections of density from the composite cryo-EM map are represented by a semi-transparent molecular surface, shaded in grey. Please also see associated key for additional detail.