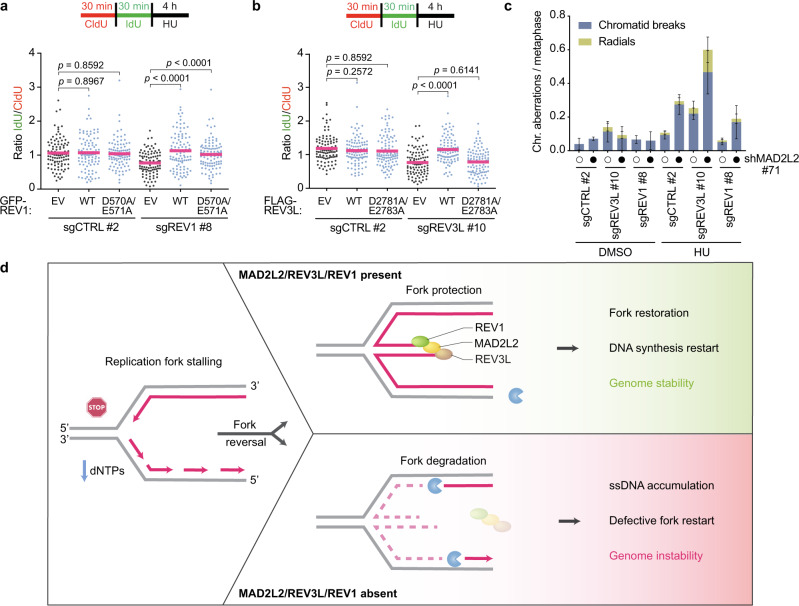
Fig. 7. MAD2L2, REV1 and REV3L polymerase activity are required for replication fork protection and genome maintenance.
a Schematic and quantification of fork degradation assays in control (sgCTRL) and REV1-depleted HeLa cells complemented with either empty vector (EV), GFP-tagged wild-type REV1 (WT), or GFP-tagged catalytic inactive mutant REV1 (D570A/E571A). A representative fiber experiment from three independent biological replicates is shown. b Schematic and quantification of fork degradation assays in control and REV3L-depleted HeLa cells complemented with either empty vector (EV), FLAG-tagged wild-type REV3L (WT), or FLAG-tagged catalytic inactive mutant REV3L (D2781A/E2783A). A representative fiber experiment from three independent biological replicates is shown. Pink bars in fiber plots represent the mean. Statistical analysis in (a, b) was performed according to two-tailed Mann-Whitney test. Additional replicates and combined fiber plots are provided in the Supplementary Information. c Quantification of chromosomal aberrations in control, REV1- and REV3L-depleted HeLa cells transduced with a control shRNA (shSCR, open circles) or MAD2L2 shRNA (closed circles). Cells were harvested following treatment with 4 mM HU for 4 h and recovery for 24 h. Bars represent the mean ± SD of three independent biological replicates. d Schematic model depicting the role of MAD2L2/REV3L/REV1 in fork protection and genome maintenance. Upon replication fork stalling due to e.g. nucleotide depletion, remodeling of stalled forks can generate a regressed, four-stranded DNA structure that is susceptible to the action of cellular nucleases such as MRE11. MAD2L2, in collaboration with REV1 and REV3L, stabilizes and protects reversed replication forks, thereby allowing efficient restart of DNA synthesis. In the absence of MAD2L2/REV3L/REV1, reversed forks undergo extensive MRE11-dependent nucleolytic degradation, which compromises fork restart and results in ssDNA accumulation and genome instability. Created with BioRender.com.