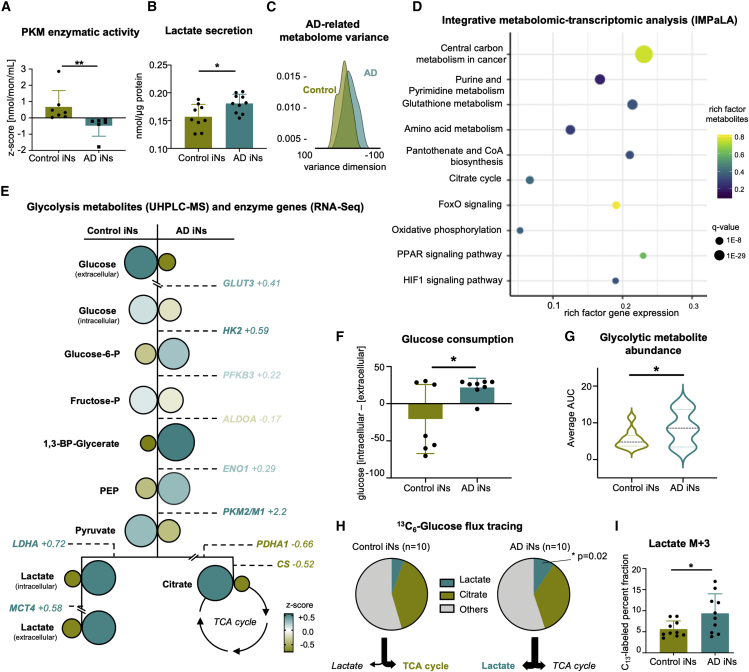
Figure 4.
Metabolome reveals Warburg-like metabolic switch in AD iNs
(A) Colorimetric assay to determine PKM activity in control (n = 7) and AD (n = 6) iNs (Mann-Whitney test).
(B) Colorimetric assay to detect secreted lactate in the supernatant of control (n = 9) and AD (n = 10) iNs.
(C) Density plot of PC7 of 160 metabolites measured by UHPLC-MS of control (n = 10) and AD (n = 10) iNs.
(D) Integrative transcriptomics and metabolomics (IMPaLa) analysis showing over-represented pathways including gene and metabolites rich factors.
(E) UHPLC-MS metabolic landscape in control (left; n = 10) and AD (right; n = 10) iNs. Circles represent Z scores of respective metabolites. Size and color of the font indicate the RNA-seq expression levels of related enzymes.
(F) Glucose consumption measured as the drop of extracellular glucose after 6 h in culture.
(G) Combined averages of all detected glycolytic metabolites in iNs.
(H and I) Tracing of isotope-labeled glucose after 6-h incubation with 13C6-glucose. Fraction of labeled glucose detected in lactate and citrate (n = 10 per group).
Dots represent individual donors throughout the figure. Bars, mean; error bars, SD; violin plots, median and quartiles. Significance: unpaired t test, ∗p < 0.05, ∗∗p<0.01.