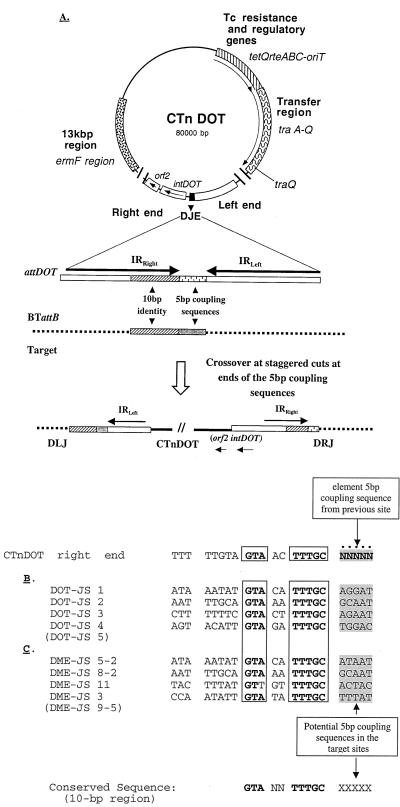
FIG. 5.
Model for the integration of CTnDOT into Bacteroides target sites. (A) Circular intermediate of CTnDOT after conjugal transfer and prior to integration with the known regions of the element indicated. The attDOT in the DJE is enlarged to show the 23-bp imperfect inverted repeats (IRRight and IRLeft), the 10 bp of high similarity to a Bacteroides target site (BTattB), and a 5-bp coupling sequence derived from its last site of integration. Details of the 10-bp alignment of the attDOT region and some attB sites are shown in panels B and C. Following staggered cuts flanking the coupling sequences in the attDOT and attB, the CTnDOT (or minielement) integrates into the target site. The 10-bp consensus sequence in the CTnDOT right end in the circular form is shown above the attB sequences in panels B and C, with the coupling sequence indicated by N's. The sequences in panel B are from the chromosomal target sites prior to integration of the circular form of the CTnDOT (DOT-JS). The sequences in panel C are from the chromosomal site prior to integration of pDJE2.3 (DME-JS). A conserved sequence GTANNTTTGC (10-bp region) was derived from a comparison of the CTnDOT right end and its target sites. The minielement target sites DME-JS 5-2 and DME-JS 8-2 are the same as the wild-type CTnDOT sites DOT-JS 1 and DOT-JS 2, respectively. After conjugal transfer and integration of pDJE2.3 (not shown), the coupling sequence from pDJE2.3 was found at the left side of the integrated element, adjacent to the 10-bp attB sequence, in DME-JS 3, 5-2, and 9-5 and the right side of the integrated pDJE2.3 in DME-JS 8-2 and 11 (sequence not shown). Parentheses indicate that the site was the same as the site above it.