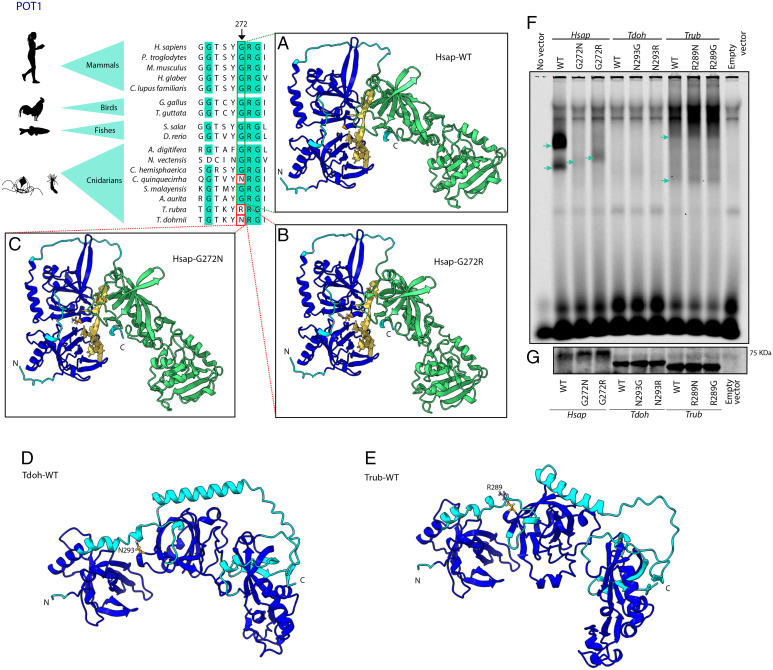
Fig. 3.
Structural and functional comparisons between POT1 from Turritopsis and other species. The figure shows a partial amino acid sequence alignment of POT1 from Turritopsis and other cnidarians, invertebrates and vertebrate species. Significant variants in T. dohrnii and T. rubra are highlighted with a red box and the arrow indicates the specific position. The figure also shows protein structures built with AlphaFold using H. sapiens structure (A) wild-type, (B) with T. rubra, and (C) T. dohrnii variants, and using (D) T. dohrnii and (E) T. rubra wild-types. Dark blue surface indicates oligonucleotide binding fold (OBF) domain which binds to ssDNA and green area represents OB-fold domain and a Holliday junction resolvase (HJR) domain which make dimer contacts with TPP1. Yellow structure is a tentative representation of the telomeric region, located by superimposing POT1-telomere model (PDB ID 1XJV) on each AlphaFold model. (F) Binding of the different POT1 variants to a single-stranded oligonucleotide containing three telomeric repeats. Blue arrows point to POT1-(TTAGGG)3 complexes (G) Western blot to check the amount of POT1 loaded in the binding assay.