Abstract
Azospirillum species are plant-associated diazotrophs of the alpha subclass of Proteobacteria. The genomes of five of the six Azospirillum species were analyzed by pulsed-field gel electrophoresis. All strains possessed several megareplicons, some probably linear, and 16S ribosomal DNA hybridization indicated multiple chromosomes in genomes ranging in size from 4.8 to 9.7 Mbp. The nifHDK operon was identified in the largest replicon.
The genome organization among the alpha subclass of Proteobacteria (α-Proteobacteria) is highly variable and complex (17). Among the Rhizobiaceae, the Bradyrhizobium japonicum genome consists of a single circular chromosome of ca. 8.7 Mbp and two plasmids (0.2 and 0.8 Mpb) (13, 17), whereas Rhizobium galegae and Rhizobium fredii have one circular chromosome and two megaplasmids and Rhizobium leguminosarum has one circular chromosome, one megaplasmid, and two small plasmids (10). On the other hand, Agrobacterium tumefaciens strain C58 has four replicons, including two DNA molecules of high molecular weight, one of which is linear and both of which hybridized with a 16S ribosomal DNA (rDNA) probe (1), suggesting that this organism possesses two chromosomes. Other genera show two chromosomes and two plasmids, as in Ochrobactrum (10) and Sinorhizobium meliloti (17). The presence of high-molecular-weight forms of DNA carrying 16S rDNA has also been shown in Brucella (9), Rhodobacter (8), rickettsiae (25), Mycoplana (10), and Phyllobacterium (10, 17). The sizes of these putative chromosomes and plasmids in the alpha subclass vary among strains (10). These results challenge the idea that prokaryotic genomes consist of a single circular chromosome and also suggest that bacterial genomes are dynamic entities and may have evolved by exchanging genetic information.
Azospirillum spp. are diazotrophs associated with several plants, including wheat and maize (7), and they are classified within the alpha subclass of the Proteobacteria by 16S rRNA sequence analysis (32). The genus comprises six species, A. brasilense, A. lipoferum (30), A. amazonense (15), A. irakense (11), A. halopraeferens (23) and A. largimobile (28). Although the benefit from biological nitrogen fixation is disputed, association of gramineae with A. brasilense or A. lipoferum has been reported to result in a more robust root system, increasing absorption of water and minerals from the soil, and faster plant growth (2, 18). A. brasilense and A. lipoferum contain several plasmids with sizes ranging from 40 kbp to 550 kbp, none of which hybridized with a probe containing nif genes (22, 31). The 90-MDa plasmid of A. brasilense strain Sp7 has been mapped by restriction enzymes and DNA hybridization, and five loci have been identified: nodH, nodN, exoB, exoD, and its probable origin of replication (19, 21, 22).
Despite several reports describing the presence of plasmids in A. brasilense and A. lipoferum (7, 19, 21, 22, 31), information about their genome size is imprecise to date. In 1982, Wood et al. (31), using a modified Eckhardt electrophoresis method, described the presence of several very large DNA bands with molecular sizes up to 2.8 Mbp that they called minichromosomes, based on their apparent large molecular size. They estimated the Azospirillum genome size as 1.8 times larger than that of Escherichia coli.
In this paper, we describe the presence of several megareplicons in 10 strains of five Azospirillum species with molecular sizes ranging from 0.2 to 2.7 Mbp as determined by pulsed-field gel electrophoresis (PFGE). The PFGE DNA patterns differ within the same species, which indicates that they are strain specific. In all strains tested, the presence of 16S rDNA was detected in more than one replicon, suggesting that Azospirillum contains multiple chromosomes. Also, the PFGE behavior indicates that some of the replicons are probably linear DNA molecules.
The Azospirillum species analyzed were A. brasilense, strains Sp7 (ATCC 29145) (30), Cd (ATCC 29710) (29), FP2 (20), and Sp245 (2); A. lipoferum, strains Sp59b (ATCC 29707) (30) and JA25 (5); A. amazonense, strains Y2 (ATCC 35120) and Y6 (ATCC 35121) (16); A. irakense (11); and A. halopraeferens (23). All bacterial strains were grown in NFbHP-malate (12) or DYGS medium (2 g of glucose/liter, 2 g of malic acid/liter, 1.5 g of peptone/liter, 2 g of yeast extract/liter, 0.5 g of K2HPO4/liter, 0.5 g of MgSO4 · 7H2O/liter, 1.5 g of glutamic acid/liter [pH 6.8]) at 30°C in a rotary shaker, except for A. irakense and A. halopraeferens, which were grown at 37°C.
The intact genome of Azospirillum was analyzed by PFGE (27). The cells were grown in liquid medium to an optical density at 600 nm ranging from 0.2 to 0.7, depending on the strain, and chromosomal DNA was purified as described previously (14) and analyzed in agarose gels (1.2%) using a Gene Navigator pulsed-field system (Pharmacia).
Bacterial cells were embedded in 100 μl of low-melting-point agarose in buffer SET (50 mM Tris-HCl [pH 7.5], 20 mM EDTA, 200 mM NaCl). Lysis was achieved by using a lysis solution (10 mM Tris [pH 7.5], 50 mM NaCl, 100 mM EDTA, 0.2% deoxycholate, 0.5% N-lauryl sarcosine) with lysozyme (1 mg/ml) at 37°C for 24 h. Protein digestion was carried out using proteinase K (0.1 mg/ml) in 0.5 mM EDTA (pH 8.0) and 1% N-lauryl sarcosine at 52°C for 48 h.
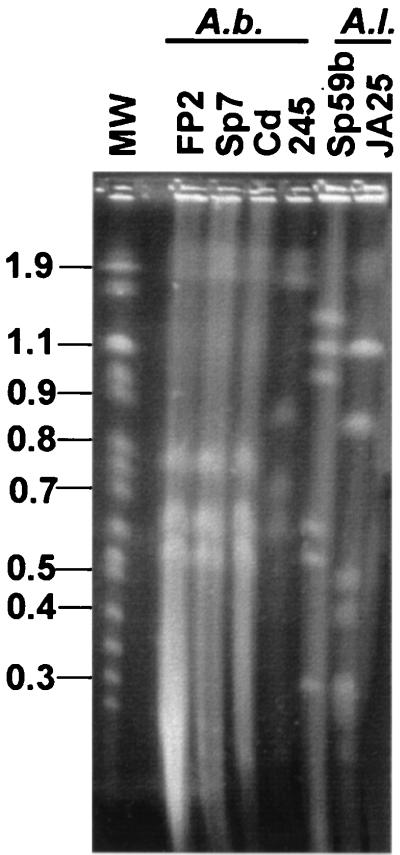
The A. brasilense strains analyzed showed five megareplicons, ranging in size from 0.63 to 2.5 Mbp (Fig. 1 and Table 1). The strains FP2, Sp7, and Cd showed the same DNA profile for that range, a result consistent with FP2 and Cd being related to Sp7. The strain FP2 is a spontaneous Sp7 mutant resistant to both nalidixic acid and streptomycin (20), and Cd was isolated from Digitalia after inoculation with Sp7 (29). These results indicate that DNA replicons of A. brasilense are stable even after intense manipulations and reisolation of the bacteria. The A. brasilense strain Sp245 also showed five megareplicons; however, the DNA profile was clearly different: the two largest bands apparently comigrate with those of Sp7, suggesting similarity, but the three smaller megareplicons showed sizes different from those for Sp7.
FIG. 1.
PFGE profile of intact genomic DNA of Azospirillum spp. The indicated strains of A. brasilense (A.b.) and A. lipoferum (A.l.) were analyzed by PFGE, with the following parameters: pulses of 60 s for 15 h and of 120 s for 9 h at 200 V, using a Gene Navigator system (Amersham Pharmacia). The chromosomes of Saccharomyces cerevisiae (Amersham Pharmacia) were used as molecular size markers (MW), and the numbers at left represent mega-base pairs.
TABLE 1.
Molecular size of replicons of Azospirillum spp., as determined by PFGE
| Species | Strain | Sizes (Mbp) ofc:
|
|
|---|---|---|---|
| Repliconsd | Genome (estimated) | ||
| A. brasilense | FP2 | 2.5ab; 1.72a; 0.81a (L); 0.7 (L); 0.63a (L); 0.17; 0.15 | 6.7 |
| Sp7 | 2.5ab; 1.74a; 0.81a (L); 0.70 (L); 0.64a (L); 0.21; 0.2 | 6.8 | |
| Cd | 2.6a; 1.77a; 0.81a (L); 0.71 (L); 0.64a (L); 0.21; 0.19 | 6.9 | |
| Sp245 | 2.6a; 1.76a; 0.9a (L); 0.78 (L); 0.72a (L); 0.21; 0.14 | 7.1 | |
| A. lipoferum | Sp59b | 2.6a; 1.8a; 1.38a; 1.18a (L); 0.97a (L); 0.71 (L); 0.65a (L); 0.4 | 9.7 |
| JA25 | 2.25(ND); 1.8a; 1.1a (L); 0.85a (L); 0.55a (L); 0.45 (L); 0.3; 0.27; 0.22; 0.15 | 7.9 | |
| A. amazonense | Y2 | 2.7a; 2.2; 1.7a; 0.75 | 7.3 |
| Y6 | 2.6a; 2.1; 1.8a; 0.71 | 7.2 | |
| A. irakense | 2.4a, 1.2a, 0.95a; 0.22 | 4.8 | |
| A. halopraeferens | 2.6a; 1.2a; 0.98a; 0.92a; 0.22 | 5.9 | |
Hybridized with 16S rDNA.
Hybridized with nifHDK.
The indicated molecular sizes are the averages of at least five determinations (except for those of A. irakense and A. halopraeferens, for which two independent experiments were performed), with a standard deviation of less than 10%. Chromosomes of S. cerevisiae, Schizosaccharomyces pombe, or λ DNA concatemers (Amersham Pharmacia or Bio-Rad) were used as molecular size markers. The genome sizes were estimated based on those of the indicated replicons.
Abbreviations: (L), Indication of linear molecule; ND, hybridization with 16S rDNA not determined.
The PFGE DNA profiles of A. lipoferum Sp59b and JA25 showed 8 and 10 replicons, respectively, with molecular sizes ranging from 0.15 to 2.6 Mbp, and none of the replicons seemed to comigrate (Fig. 1 and 2 and Table 1). Both A. amazonense strains, Y2 and Y6, showed four replicons but had distinct PFGE profiles, with sizes varying from 0.71 Mbp (Y6) to 2.8 Mbp (Y2). Several replicons were also observed in A. halopraeferens and A. irakense (Fig. 2 and Table 1), each strain with a specific DNA pattern. This is the first report of the presence of megareplicons in A. amazonense, A. irakense, and A. halopraeferens.
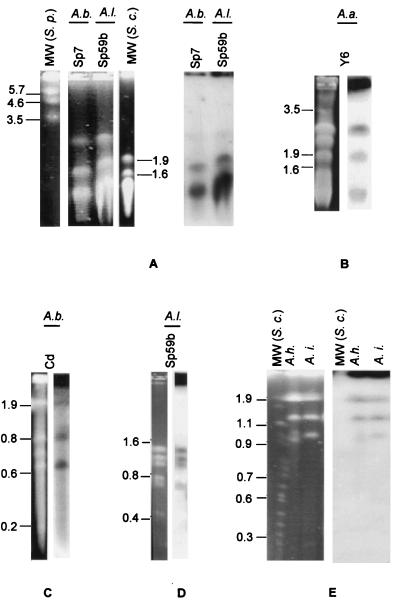
FIG. 2.
Identification of 16S rDNA in Azospirillum replicons by hybridization. A PFGE profile of intact DNA of the indicated strains (left) and an autoradiogram of hybridization with the 32P-labeled A. brasilense 16S rDNA gene, performed as described by Sambrook et al. (26) (right), are shown. (A) A. brasilense strain Sp7 and A. lipoferum strain Sp59b, analyzed with a pulse gradient of 10 to 50 min for 120 h at 55 V. (B) A. amazonense strain Y6 with a similar pulse gradient for 90 h. (C) A. brasilense strain Cd, analyzed using pulses of 60 s for 15 h and 90 s for 9 h at 200 V. (D) A. lipoferum strains Sp59b, analyzed using a gradient of 1 to 10 s for 36 h, following a gradient of 1 to 2 min for 12 h at 120 V. (E) A. halopraeferens (A.h.) and A. irakense (A.i.), analyzed using PFGE parameters of 60 s for 17 h and 120 s for 7 h at 200 V. The chromosomes of S. cerevisiae (S.c.) (Amersham Pharmacia) and S. pombe (Bio-Rad) were used as molecular size markers (MW), and the numbers at left represent mega-base pairs. PFGE was performed using a Gene Navigator system (Amersham Pharmacia).
The overall genome size of members of the genus Azospirillum varied from a minimum of 4.8 Mbp (A. irakense) to 9.7 Mbp (A. lipoferum strain Sp59b). These results indicate that the organization of the Azospirillum genome is highly complex, with the genetic information distributed on several replicons. In addition, the DNA pattern was strain specific rather than species specific, a result also observed for Brucella (9). The role of these replicons in the ecological and in vitro survival of these species remains to be determined but may support the exceptional ecological distribution and metabolic flexibility of members of this genus (9).
According to Römling et al. (24), only linear DNA molecules permeate a gel and can be separated by PFGE. Large circular DNA molecules do not permeate the gel and can be analyzed only after linearization either by physical or enzymatic treatment or randomly during DNA preparation. Because the bacterial lysis conditions are made very mild to avoid DNA breakage, randomly broken DNA produced from circular molecules is in low concentration and therefore shows weak, less intense bands, which also vary with the method of preparation, on a PFGE gel. The two largest replicons of A. brasilense and A. lipoferum showed less intensity than the others bands (Fig. 1 and 2), and their relative intensities varied with the method of preparation (data not shown), suggesting that those replicons were circular DNA molecules. In addition, partial DNA digestions of strains FP2 and JA25 with very low concentrations of restriction enzymes to produce a single cut per molecule caused an increase in the intensities of those bands relative to the others, confirming that they were produced from circular DNA (data not shown). A different behavior was observed, however, with the other replicons when intact genomic DNA was analyzed by PFGE. A. brasilense and A. lipoferum showed very intense bands of molecular sizes ranging from 0.4 to 1.1 Mbp (Fig. 1) that behaved like linear DNA, with no variation in the relative intensities of the bands after partial endonucleolytic digestion (not shown). These results strongly suggest that the A. brasilense and A. lipoferum genomes contain both linear and circular DNA. In A. amazonense, replicons of 0.75 (Y2) and 0.71 (Y6) Mbp were considered to be circular DNA. The same topology was observed for the 0.22-Mbp replicon found in A. irakense and A. halopraeferens. More information, however, is necessary to determine whether some of the replicons in A. amazonense, A. irakense, and A. halopraeferens were linear DNA molecules, although several of them showed very intense bands in the PFGE of intact DNA.
Due to the high molecular weight of Azospirillum replicons, we reasoned that essential genes might be present in more than one replicon, indicating the presence of multiple chromosomes. DNA hybridization studies showed that all strains tested had at least two replicons hybridizing with a 16S rDNA probe from A. brasilense (Fig. 2 and Table 1). Wood et al. (31) reported the presence of minichromosomes in A. brasilense and A. lipoferum, based on the sizes of the DNA bands determined by vertical Eckhardt-type agarose gel electrophoresis; however, no localization or mapping of essential genes was reported. The DNA profile of Sp7 obtained by Wood et al. (31) was very similar to ours, with three bands in the region of 0.6 Mbp and two bands above 1.7 Mbp. Recently, Caballero-Mellado et al. (4) also analyzed several strains of A. brasilense by a horizontal Eckhardt-type gel electrophoresis and reported the presence of more than one replicon carrying 16S rDNA genes. However, these authors did not observe DNA hybridization with the largest replicon of strain Sp7 and suggested that this was probably due to its very high molecular weight (4). In addition, the genome profile of the Sp7 strain obtained by Caballero-Mellado et al. (4) was slightly different from that reported here and from that obtained by Wood et al. (31), a result probably due to electrophoresis resolution of the different experimental conditions.
While the 16S rRNA gene was found in several replicons in Azospirillum, the nifHDK operon seems to be located in only one replicon, at least for A. brasilense strains FP2 and Sp7. These genes were present in the largest replicon of these strains (Table 1), as revealed by DNA hybridization with an A. brasilense nifHDK probe. In the other strains, the nifDK genes were clearly located in a circular DNA molecule, since a very intense hybridization signal was observed in the gel wells in a PFGE of intact DNA (data not shown).
Azospirillum spp. have one of the most complex patterns of high-molecular-weight DNA among the α-Proteobacteria so far described. All five species analyzed showed multiple replicons, and the presence of 16S rDNA genes in several of them supports the suggestion that the Azospirillum genome is split into several chromosome-like structures. In A. brasilense and A. lipoferum, these structures were found in either linear or circular DNA, as has also been observed for other species within the alpha subclass of the Proteobacteria (17).
The differences in the DNA patterns found among strains of the same species suggest that although stable, the genome structures of these organisms seem to evolve faster than the species differentiation. The mechanism underlying the development of these genome structures is unknown but probably involves genetic rearrangements between homologous DNA sequences shared by two or more replicons, as shown in Brucella (10). It may also involve horizontal DNA exchange.
Acknowledgments
We are grateful to Roseli Prado, Valter A. Baura, and Julieta Pie for technical assistance. We also thank EMBRAPA-Agrobiologia (Seropédica, RJ, Brazil) and EMBRAPA-Trigo (Passo Fundo, RS, Brazil) for the Azospirillum strains. L.S.C. thanks Meire C. Delgado-Bremer for helpful technical information about PFGE.
This work was supported by CNPq and PRONEX (FINEP/MCT/CNPq).
REFERENCES
- 1.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1998;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldani V L D, Baldani J I, Dobereiner J. Inoculation of field-grown wheat (Triticum aestivum) with Azospirillum spp. in Brazil. Biol Fertil Soils. 1987;4:37–40. [Google Scholar]
- 3.Burdman S, Kigel J, Okon Y. Effects of Azospirillum brasilense on nodulation and growth of common bean (Phaseolus vulgaris L.) Soil Biol Biochem. 1997;29:923–929. [Google Scholar]
- 4.Caballero-Mellado J, Lopez-Reyes L, Bustillos-Cristales R. Presence of 16S rRNA genes in multiple replicons in Azospirillum brasilense. FEMS Microbiol Lett. 1999;178:283–288. [Google Scholar]
- 5.Didonet A D. Ph.D. thesis. Campinas, Brazil: UNICAMP; 1993. [Google Scholar]
- 6.Döbereiner J. The genera Azospirillum and Herbaspirillum. In: Ballows A, Trupper H G, Dworking M, Harder W, editors. The procaryotes. 2nd ed. III. Berlin, Germany: Springer-Verlag KG; 1991. p. 2236. [Google Scholar]
- 7.Döbereiner J, Pedrosa F O. Nitrogen-fixing bacteria in nonleguminous crop plants. Madison, Wis: Science Tech Publishers; 1987. [Google Scholar]
- 8.Fonstein M, Zheng S, Haselkorn R. Physical map of the genome of Rhodobacter capsulatus SB 1003. J Bacteriol. 1992;174:4070–4077. doi: 10.1128/jb.174.12.4070-4077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jumas-Bilak E, Michaux-Charachon S, Bourg G, O'Callaghan D, Ramuz M. Differences in chromosome number and genome rearrangement in the genus Brucella. Mol Microbiol. 1998;27:99–106. doi: 10.1046/j.1365-2958.1998.00661.x. [DOI] [PubMed] [Google Scholar]
- 10.Jumas-Bilak E, Michaux-Charachon S, Bourg G, Ramuz M, Allardet-Servent A. Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol. 1998;180:2749–2755. doi: 10.1128/jb.180.10.2749-2755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khammas K M, Ageron E, Grimont P A D, Kaiser P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res Microbiol. 1989;140:679–693. doi: 10.1016/0923-2508(89)90199-x. [DOI] [PubMed] [Google Scholar]
- 12.Klassen G, Pedrosa F O, Souza E M, Funayama S, Rigo L U. Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae. Can J Microbiol. 1997;43:887–891. [Google Scholar]
- 13.Kundig C, Hennecke H, Gottfert M. Correlated physical and genetic map of Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J S, Birren B, Lai E. Introduction to pulsed-field gels and preparation and analysis of large DNA. In: Birren B, Lai E, editors. Nonmammalian genomic analysis—a practical guide. San Diego, Calif: Academic Press; 1996. p. 1. [Google Scholar]
- 15.Magalhães F M M, Baldani J I, Souto S M, Kuykendall J R, Döbereiner J. A new acid-tolerant Azospirillum species. An Acad Bras Ciênc. 1983;55:417–430. [Google Scholar]
- 16.Martinez-Drets G, Fabiano E, Cardona A. Carbohydrate catabolism in Azospirillum amazonense. Appl Environ Microbiol. 1985;50:183–185. doi: 10.1128/aem.50.1.183-185.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno E. Genome evolution within the alpha Proteobacteria: why do some bacteria not possess plasmids and others exhibit more than one different chromosome? FEMS Microbiol Rev. 1998;22:255–275. doi: 10.1111/j.1574-6976.1998.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 18.Okon Y. Azospirillum as a potential inoculant for agriculture. Trends Biotechnol. 1985;3:223–228. [Google Scholar]
- 19.Onyeocha I, Vieille C, Zimmer W, Baca B E, Flores M, Palacios R, Elmerich C. Physical map and properties of a 90-MDa plasmid of Azospirillum brasilense Sp7. Plasmid. 1990;23:169–182. doi: 10.1016/0147-619x(90)90049-i. [DOI] [PubMed] [Google Scholar]
- 20.Pedrosa F O, Yates M G. Regulation of nitrogen fixation (nif) genes of Azospirillum brasilense by nifA and ntrC (glnG) type genes. FEMS Microbiol Lett. 1984;55:95–101. [Google Scholar]
- 21.Penot I, Berges N, Guinguene C, Fages J. Characterization of Azospirillum associated with maize (Zea Mays) in France using biochemical tests and plasmid profiles. Can J Microbiol. 1992;38:798–903. [Google Scholar]
- 22.Plazinski J, Dart P J, Rolfe B G. Plasmid visualization and nif gene location in nitrogen-fixing Azospirillum strains. J Bacteriol. 1983;155:1429–1433. doi: 10.1128/jb.155.3.1429-1433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinhold B, Hurek T, Fendrik I, Pot B, Gillis M, Kesters K, Thielemans S, De Ley J. Azospirillum halopraeferens sp. nov., a nitrogen-fixing organism associated with the roots of Kallar grass (Leptochloa fusca (L.) Kunth) Int J Syst Bacteriol. 1987;37:43–51. [Google Scholar]
- 24.Römling U, Fislage R, Tümmler B. Macrorestriction mapping and analysis of bacterial genomes. In: Birren B, Lai E, editors. Nonmammalian genomic analysis—a practical guide. San Diego, Calif: Academic Press; 1996. p. 165. [Google Scholar]
- 25.Roux V, Raoult D. Genotypic identification and phylogenetic analysis of the spotted fever group rickettsiae by pulsed-field gel electrophoresis. J Bacteriol. 1993;175:4895–4904. doi: 10.1128/jb.175.15.4895-4904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 28.Sly L I, Stackebrandt E. Description of Skermanella parooensis gen. nov., sp. nov. to accommodate Conglomeromonas largomobilis subsp. largomobilis to the genus Azospirillum. Int J Syst Bacteriol. 1999;49:541–544. [Google Scholar]
- 29.Tal S, Okon Y. Production of the material poly-β-hydroxybutyrate and its function in Azospirillum brasilense Cd. Can J Microbiol. 1985;31:608–613. [Google Scholar]
- 30.Tarrant J J, Krieg N R, Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerink) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978;24:967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- 31.Wood A G, Menezes E M, Dykstra C, Duggan D E. Methods to demonstrate the megaplasmids (or minichromosomes) in Azospirillum. In: Klingmüller W, editor. Azospirillum I: genetics, physiology, ecology. Basel, Switzerland: Burkhäuser Verlag; 1982. p. 18. [Google Scholar]
- 32.Young J P W. Phylogenetic classification of nitrogen-fixing organisms. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1992. p. 43. [Google Scholar]