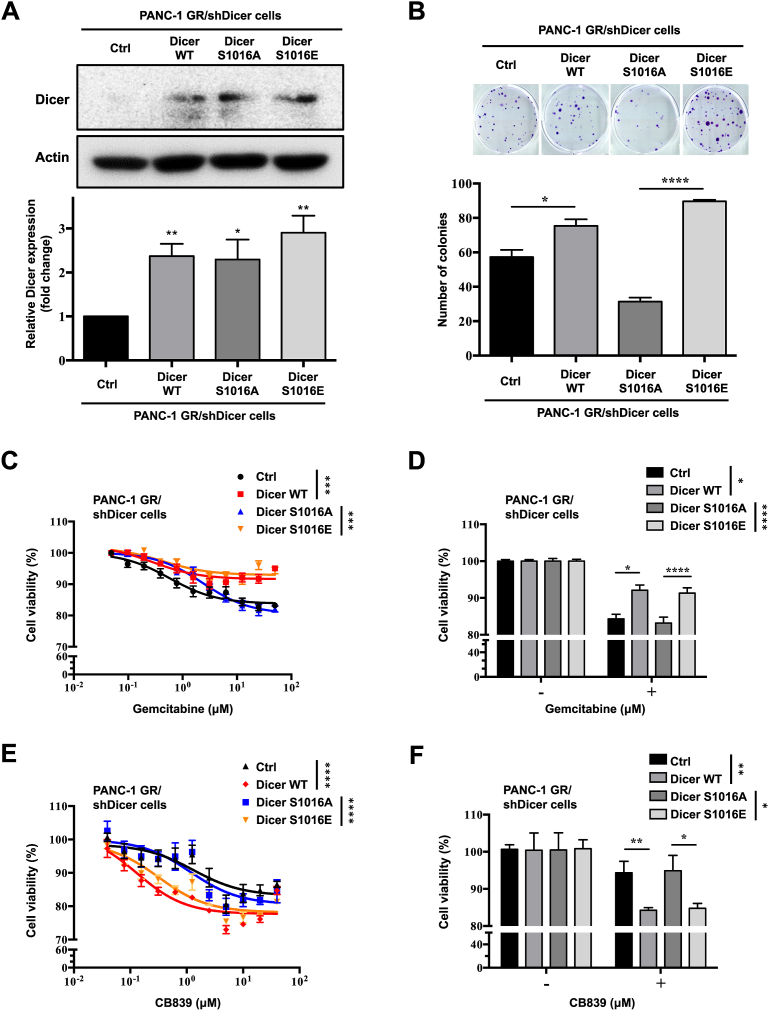
Figure 5.
Phosphomimetic Dicer S1016E in PANC-1 GR/shDicer cells increasecell viability, GEM resistance, and GLS inhibitor response. (A) Upper panel: Dicer protein expression in phosphomimetic Dicer S1016E (Dicer S1016E), phosphomutant Dicer S1016A (Dicer S1016A), Dicer WT, and control (Ctrl) PANC-1 GR/shDicer cells was analyzed through Western blotting. β-Actin was used as a loading control. Bottom panel: The relative quantification of Dicer expression in the indicated cells was performed using ImageJ software. Western blotting data were normalized to the β-actin level in each individual sample, and a bar plot presents fold changes in control cells. The results are presented as the means ± SEMs of three independent experiments. ∗P < 0.05, ∗∗P < 0.01, two-tailed Student's t-test. (B) Upper panel: colony formation of indicated cells cultured for 2 weeks; the colonies were stained with crystal violet and counted using ImageJ software. Bottom panel: Data are expressed as the number of colonies, and results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05 and ∗∗∗∗P < 0.0001, one-way ANOVA. (C) The indicated cells were incubated with varying doses of GEM for 48 h, and cell viability was analyzed using the MTT assay. (D) Bar plot comparing results between the untreated (−, 0 μM) and treated (+, 2.5 μM) groups. (E) The indicated cells were incubated with varying doses of the GLS inhibitor CB839 for 72 h, and cell viability was analyzed using the MTT assay. (F) Bar plot comparing results between the untreated (−, 0 μM) and CB839-treated (+, 0.625 μM) groups. Results are presented as the means ± SEMs of the three independent experiments. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001, one-way ANOVA.