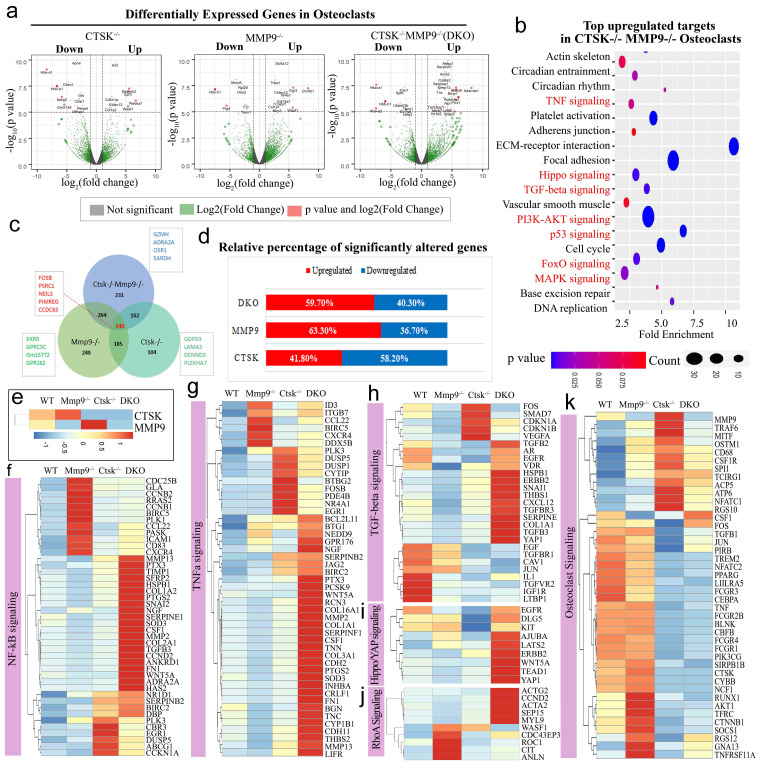
Figure 4.
RNA-seq analysis of Ctsk-/-, Mmp9-/-, and Ctsk-/-/Mmp9-/- osteoclasts showed their distinct functions in regulating genes' expression and signaling pathway activation. (a) A volcano plot illustrating differentially regulated gene expression from RNA-seq analysis between the control Ctsk-/-, Mmp9-/-, and Ctsk-/-/Mmp9-/- osteoclasts. Values are presented as the log2 of tag counts. (b) Top upregulated signaling pathways in DKO osteoclasts analyzed by KEGG database. (c) Venn diagram showing the total number of differentially expressed genes (P < 0.05) between Ctsk-/-/Mmp9-/-, Ctsk-/-, and Mmp9-/- mice. (d) RNA-seq comparison revealed the percentage of upregulated versus downregulated genes a total of 22669 genes expressed in respective mouse g©p. (e) Heatmap to confirm successful knockout of Ctsk and Mmp9. (f-k) Heatmaps of representative signaling pathways NF-κB, TNFα, TGF-beta, Hippo/YAP, RhoA, and osteoclast signaling in Ctsk-/-/Mmp9-/-, Ctsk-/-, and/or Mmp9-/- mice osteoclasts.