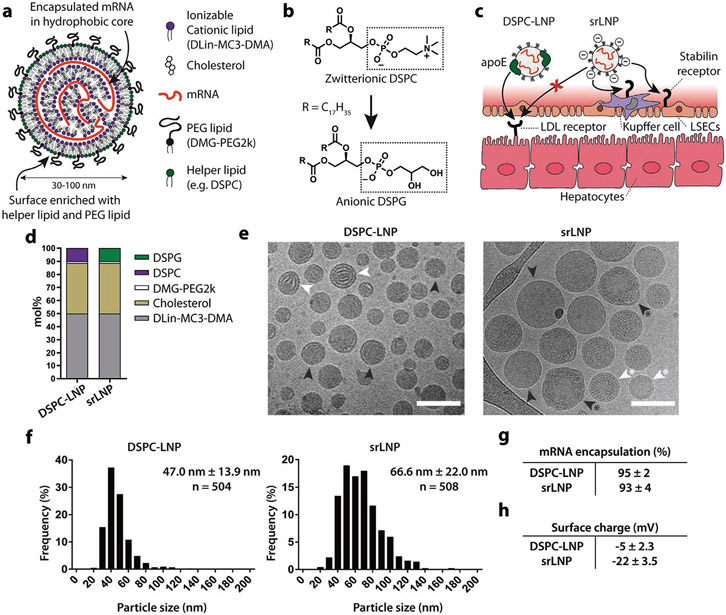
Figure 1.
Design and characterization of srLNPs. a) Schematic of the structural organization of a LNP containing mRNA, as described previously.[44] Helper phospholipids (typically incorporated at 10 mol%) are enriched at the LNP surface. b,c) Within the liver sinusoids, switching of the helper phospholipid from zwitterionic DSPC (as in Onpattro) to anionic DSPG created anionic srLNPs that are directed to the hepatic RES, via stabilin-receptor-mediated recognition and uptake in LSECs. srLNP uptake within hepatic RES cells is further enhanced by the inhibition of apoE–LDLr interactions mediated by anionic phospholipids (e.g., DSPG).[45] The mechanism(s) of recognition and uptake of srLNPs by blood resident macrophages (i.e., KCs) are not fully known. d) Lipid composition of DSPC–LNPs (i.e., Onpattro) and srLNPs. e) Cryo-EM images of DSPC–LNPs and srLNPs (entrapping capped mRNA–eCFP) showing solid lipid nanoparticle structures. Scale bars: 100 nm. Internal structures indicated with arrows: lamellar (white), amorphous (black), polymorphous (black*), and unilamellar (white*). f) Size distribution of DSPC–LNPs and srLNPs, as determined by cryo-EM. The values derived from the frequency distribution graphs represent the mean ± standard deviation (s.d.). g) mRNA encapsulation efficiency within DSPC–LNPs and srLNPs, as determined by RiboGreen assay. h) Surface charge of DSPC–LNPs and srLNPs, as determined by zeta potential measurements. See Table S1 in the Supporting Information for full biophysical characterization of all formulations used in this study.