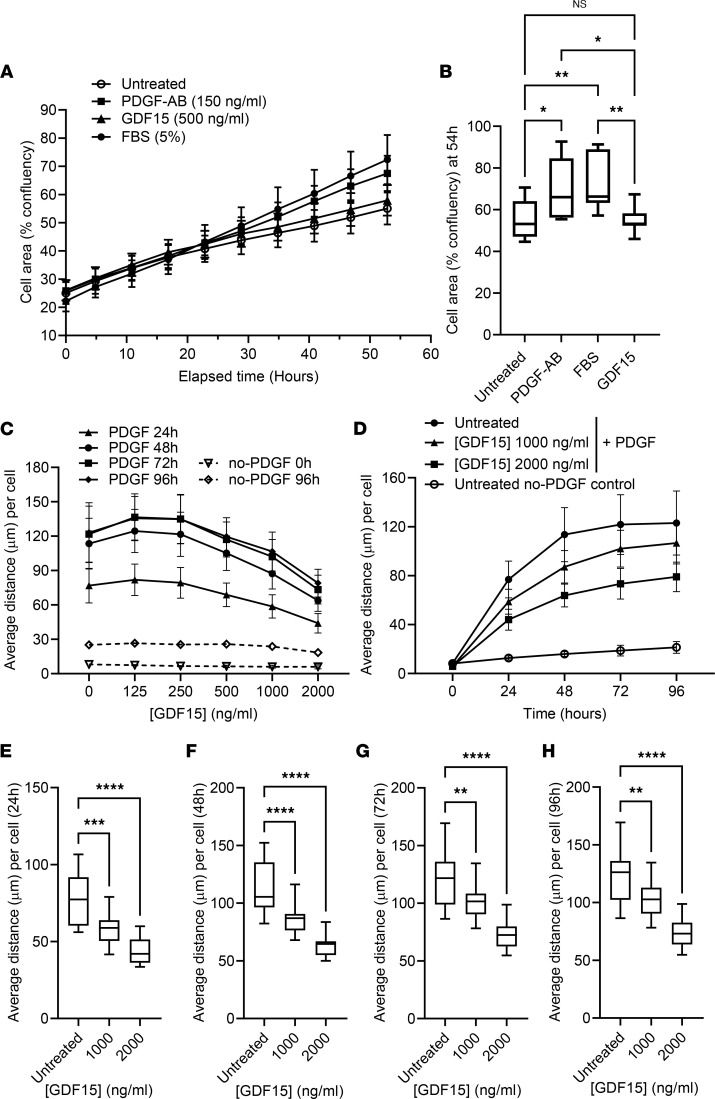
Figure 7. Stimulation with rGDF15 does not affect cell proliferation but leads to a decreased migration of NHLF.
(A and B) For assessment of cell proliferation rate, 1.6 × 103 NHLF per well were plated in the presence of rGDF15, PDGF-AB, or 5% FBS, and the cell confluency (%) was calculated over 55 hours of incubation using Incucyte system. Data are expressed as mean ± SD value over time (A) or using box-and-whisker plot (with the line in the middle plotted at the median) at 54 hours (B), statistically analyzed using multiple Kruskal-Wallis test. *P < 0.05, **P < 0.01; n = 3 biological replicates. (C) NHLF were embedded in collagen gel with or without rGDF15. After 6 hours of incubation, PDGF-BB was added to form a stable gradient, and migration of NHLF was tracked using confocal live-imaging system Yokogawa CV7000 for 96 hours. Average migration distance per cell was calculated using Columbus software. (D) PDGF-induced migration of NHLF was compared with no-PDGF, untreated control. (E–H) The data were statistically analyzed at 24 (E), 48 (F), 72 (G) and 96 (H) hours of migration toward PDGF-BB with multiple 1-way ANOVA test. **P < 0.01, ***P < 0.001 and ****P < 0.0001; n = 3 biological replicates. Data are expressed as mean ± SD (C and D) or as box-and-whisker plots (E–H), with the line in the middle plotted at the median.