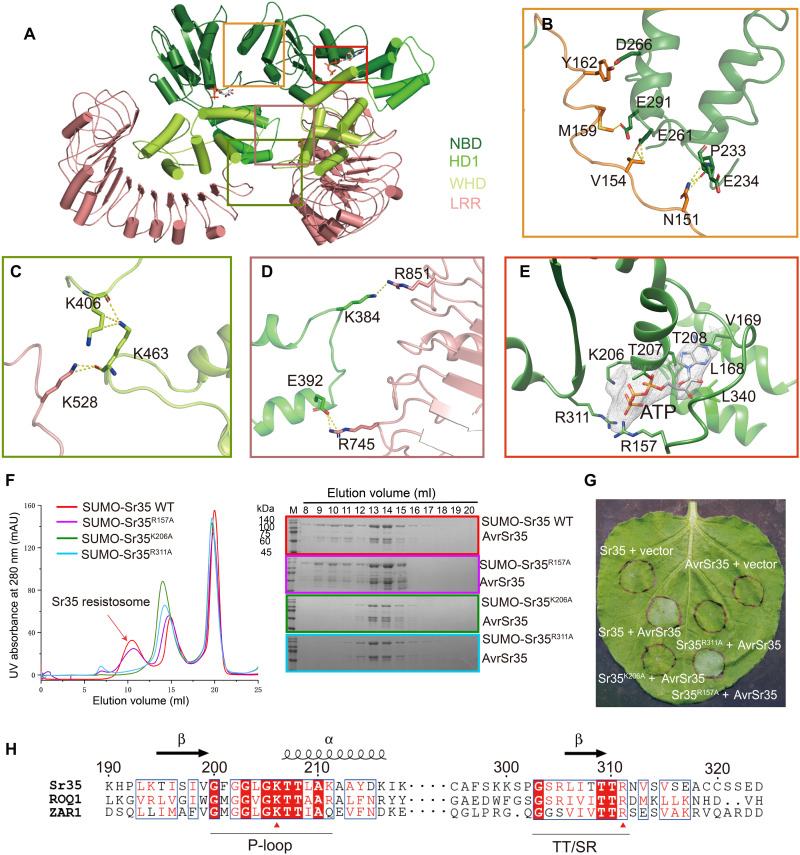
Fig. 2. Oligomerization of Sr35 resistosome.
(A) An overall view of two adjacent Sr35 protomers in the resistosome. Perspective of presentation highlight the interfaces mediating the lateral dimer. (B to D) Close-up views of the NBD-NBD, LRR-WHD, and HD1-LRR interfaces, respectively. Hydrogen bonds are shown as yellow dashed lines. (E) Interaction between ATP and Sr35. Cryo-EM density of ATP is shown as gray mesh. (F) Mutations of residues in the ATP-binding pocket affect the formation of Sr35 resistosome. Left: Gel filtration profiles of wild-type (WT) Sr35 or its mutant variants and AvrSr35 incubated in the presence of ATP. Curves are indicated in different colors, and the peak corresponding to resistosome is indicated by arrow. Right: The peak fractions in the left panel were visualized by SDS–polyacrylamide gel electrophoresis (PAGE) and Coomassie blue staining. Gels are color-coded as the left panel. UV, ultraviolet; mAU, milli–absorbance units. (G) Coinfiltration of N. benthamiana with WT and mutant Sr35 and AvrSr35 constructs. The images were taken at 36 to 48 hours postinfection (hpi). The experiment was performed three times. (H) Local sequence alignment between the Sr35, ROQ1, and ZAR1. Positions of P-loop and TT/SR motif are labeled along the sequence using underscore. Evolutionary conserved amino acids that interact with ATP are marked with a red triangle.