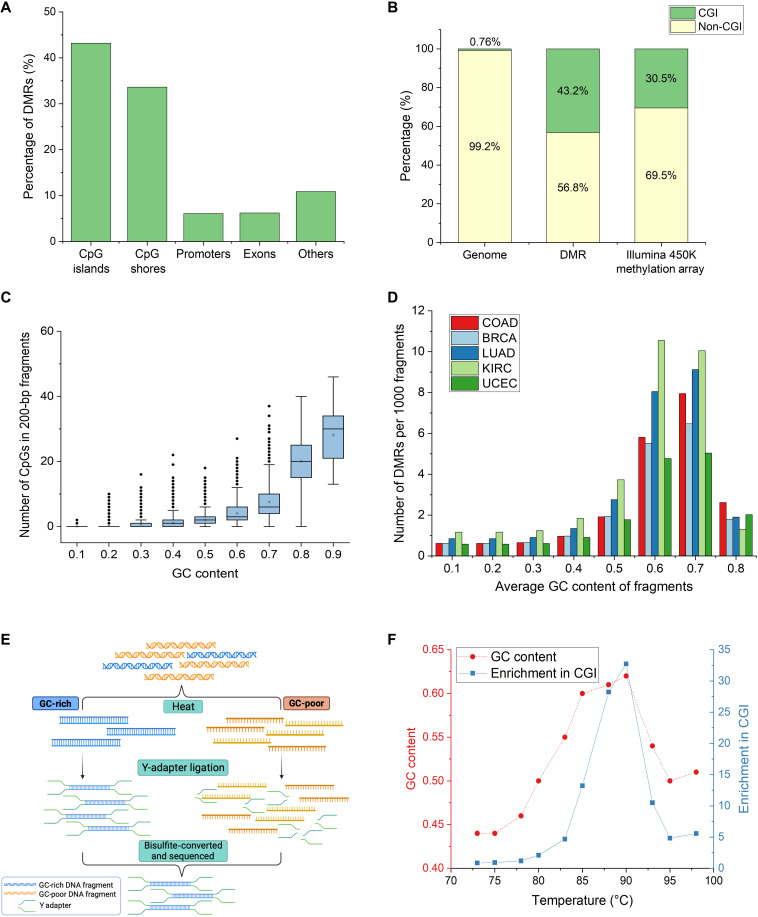
Fig. 1. CpG-rich DMRs are highly enriched in regions with high GC content.
(A) Percentage of DMRs between CRC tissue and healthy plasma in different genomic regions. More than 40% of DMRs lie in CpG-rich CGIs. (B) Proportion of DMRs and Illumina 450K Methylation Array probes in CGI with respect to the genomic distribution. Forty-three percent of DMRs and 30% of Illumina 450K methylation array probes lie in 0.76% of the genome. (C) Relationship between GC content and number of CpGs in 0.5 million randomly generated 200-bp fragments from the human genome. Fragments with high GC content also contain more CpG within each fragment. (D) Number of DMRs of different cancers detected per 1000 fragments using different GC-content thresholds. COAD, colorectal adenocarcinoma; BRCA, breast invasive carcinoma; LUAD, lung adenocarcinoma; KIRC, kidney renal clear cell carcinoma; UCEC, uterine corpus endometrial carcinoma. Fragments above 0.6 GC content contain nearly eightfold more DMRs across different cancers. (E) Workflow of Heatrich-BS to select for GC-rich fragments. GC-poor fragments are denatured by heat, and intact GC-rich fragments are selected by Y-adapter ligation. (F) Trend of GC content and read enrichment at CGI over a range of temperatures. Optimal enrichment is achieved at 88°C.