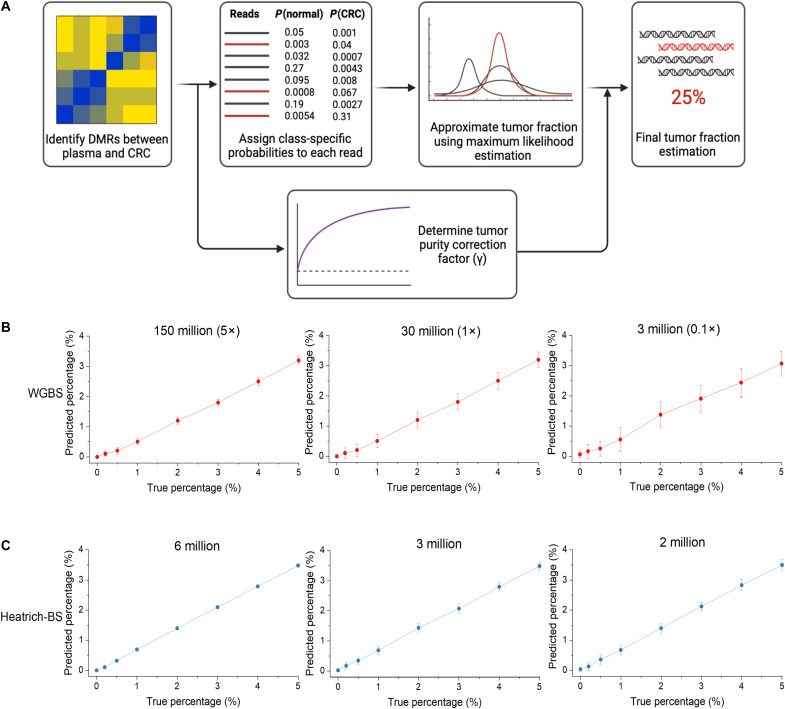
Fig. 3. Development and validation of the tumor fraction prediction algorithm.
(A) Workflow of the tumor fraction prediction algorithm. DMRs were identified by comparing healthy volunteer plasma with The Cancer Genome Atlas (TCGA) CRC methylation array data. Class-specific probabilities were assigned to each sequencing fragment, and maximum likelihood estimation was used to infer global tumor fraction. Tumor purity correction was applied to account for normal cell infiltration in TCGA data. (B) True and algorithm-predicted values of simulated plasma WGBS cfDNA samples at different sequencing depths. Confident tumor fraction prediction is achieved beyond 150 million reads (5× sequencing depth). (C) True and algorithm-predicted value of simulated Heatrich-BS samples using different total sequencing reads. Confident tumor fraction prediction is achieved with as few as 3 million reads.