Fig. 8.
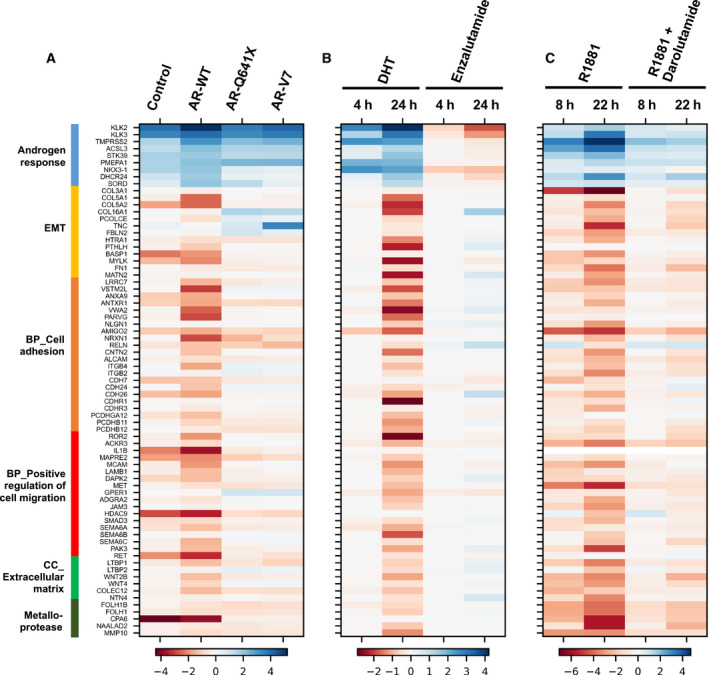
Expression profile of 73 AR‐regulated genes in GSE158557 (A), GSE125014 (B), and GSE148397 (C) data. A selection of 64 genes was used to highlight functional consequences of AR inhibition or the expression of constitutively active AR variants on epithelial‐to‐mesenchymal transition (EMT), cell adhesion and positive regulation of cell migration biological processes (BP), extracellular matrix cellular component (CC), and on metalloproteases among the three RNA‐seq datasets. In addition, nine genes known to be upregulated by AR were used as controls. DHT, dihydrotestosterone; R1881, synthetic androgen; Enzalutamide and Darolutamide, anti‐androgens.
