FIGURE 6.
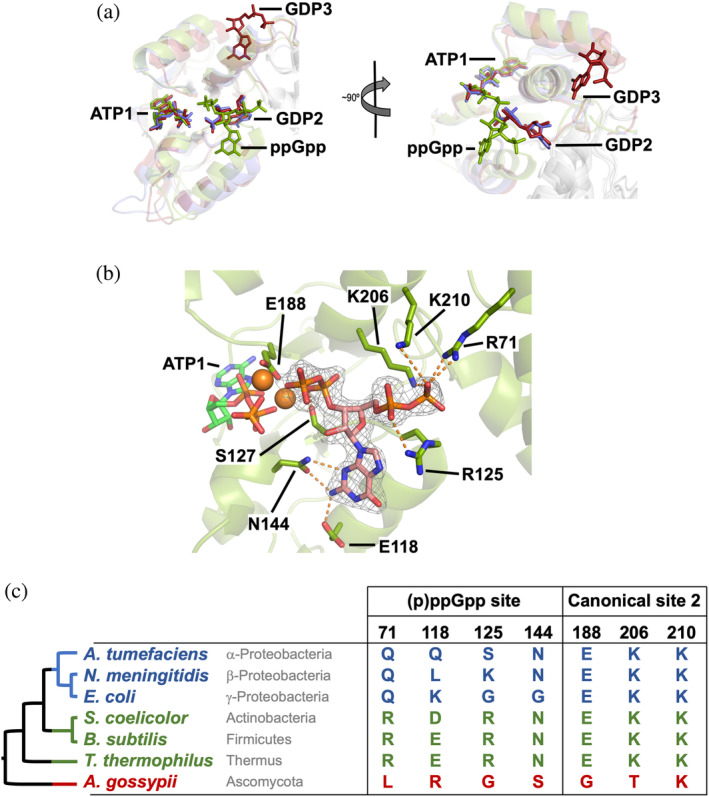
Structure of StcIMPDH bound to ATP and ppGpp. (a) Structural superimposition of the Bateman domains of PaIMPDH‐ATP/GDP (blue), StcIMPDH‐ATP‐ppGpp (green), and AgIMPDH‐ATP/GDP (red; PDB ID 5TC3). 6 (b) Detailed view of the ppGpp binding site in the Bateman domain. IMPDH protein is represented in semitransparent green cartoons with the side chain of key interacting residues shown in sticks. The 2mFo–DFc electron density map, contoured at the 1.6σ level, is shown as a grey mesh. Key protein–nucleotide atomic interactions are represented as orange dashed lines and the coordinated Magnesium atoms are shown as orange spheres. (c) The taxonomic distribution of the (p)ppGpp binding site within the Bateman domain is shown. The phylogenetic tree on the left shows the evolutionary relationships among the groups of bacteria (color‐coded according to a) and is extracted from a more detailed analysis shown in Figures S9 and S10. IMPDH, IMP dehydrogenase; PaIMPDH, Pseudomonas aeruginosa IMPDH; StcIMPDH, Streptomyces coelicolor IMPDH
