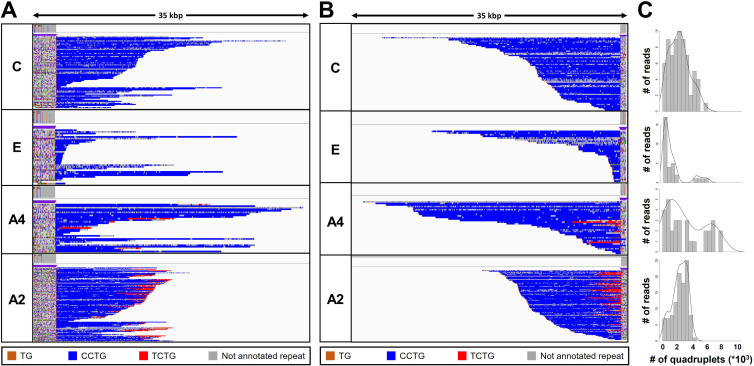
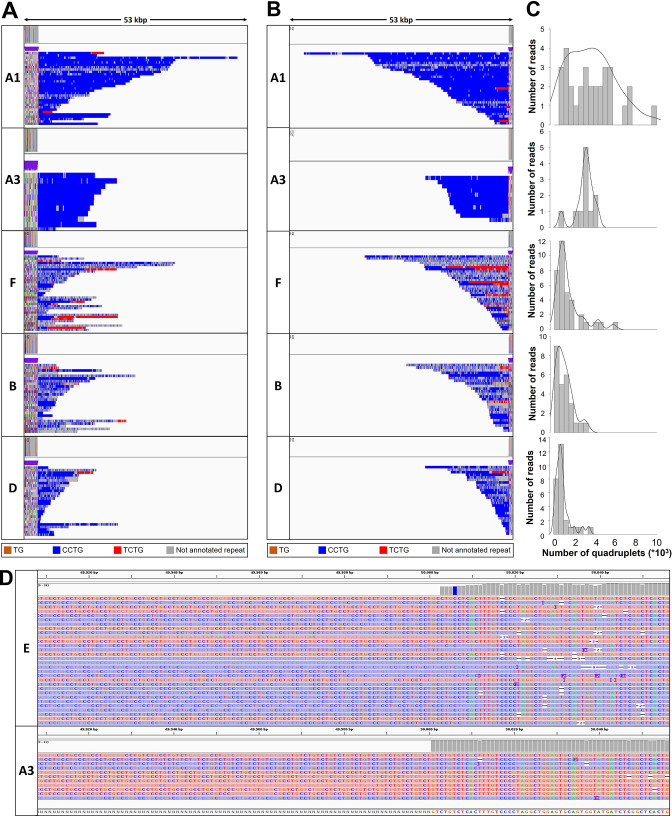
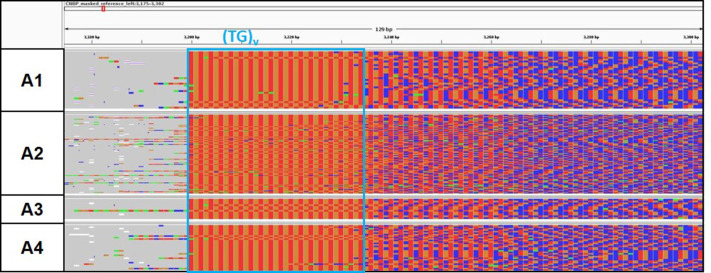
Figure 3. Analysis of the expanded-repeat CNBP alleles in dystrophy type 2 (DM2) patients.
Integrative Genomics Viewer (IGV) visualization (35-kbp windows) of ONT-targeted sequencing data from the expanded alleles of four representative DM2 patients. Complete reads were aligned at the 5′ end (A) and then at the 3′ end (B) in order to identify the repeat pattern that characterizes the expanded microsatellite locus. Each motif in the expanded alleles was visualized using a different color, as indicated in the key. Samples C and E contained a ‘pure’ CCTG expansion (blue) whereas samples A4 and A2 also contained the unexpected TCTG motif (red) downstream of CCTG. (C) Abundance of quadruplets identified in each patient. The y-axis shows the number of ONT reads with a certain number of repeats, whereas the x-axis shows the number of quadruplet repeats identified. ONT reads were grouped into 500 bp bins. The gray line represents the estimated kernel density of the underlying solid gray distribution of ONT reads.