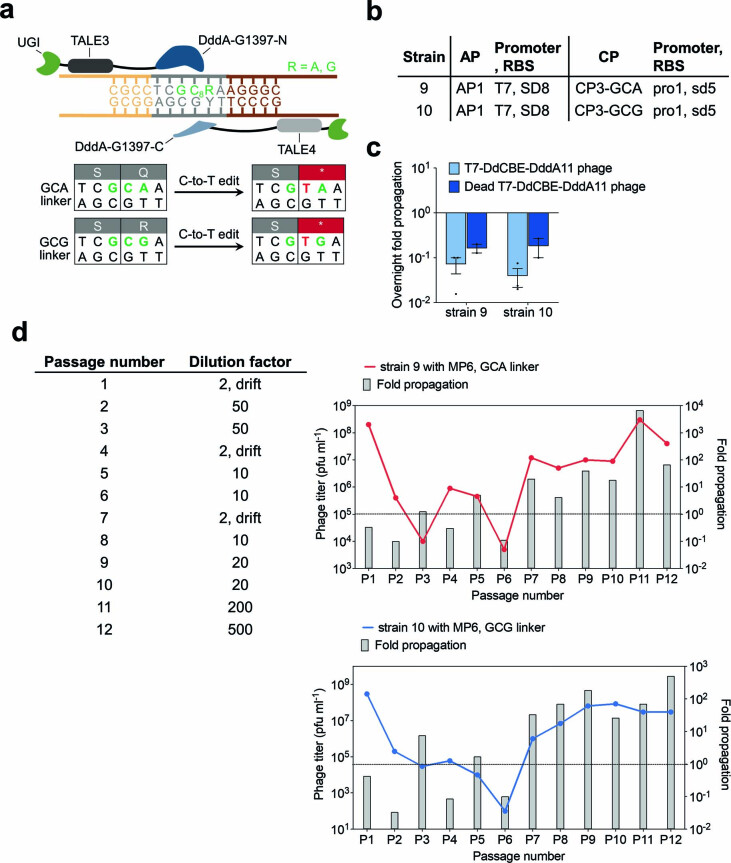
Extended Data Fig. 7. Evolution of T7-DdCBE-DddA11 using PANCE for improved GC activity.
a, The sequence encoding the T7 RNAP–degron linker was modified to contain GCA or GCG in an effort to evolve for higher activity on GC targets. T7-DdCBE must convert GC8 to GT8 to install a stop codon in the linker sequence and restore T7 RNAP activity. b, Strains for overnight phage propagation assays on GCA or GCG linkers. c, Overnight fold propagation of indicated SP in host strains encoding GCA or GCG linkers. Strains correspond to Extended Data Fig. 7b. T7-DdCBE-DddA11 phage contains the mutations S1330I, A1341V, N1342S, E1370K, T1380I and T1413I in DddA. Dead T7-DdCBE-DddA11 phage contains an additional inactivating E1347A mutation in DddA. Values and error bars reflect the mean±s.d of n = 3 independent biological replicates. d, Phage passage schedule for T7-DdCBE-DddA11 evolution in PANCE using strain 9 transformed with MP6 (red) or strain 10 transformed with MP6 (blue). The table indicates the dilution factor for the input phage population (see Methods for drifting procedure). Output phage titer and fold propagation are shown for a single replicate. Fold propagations above the dotted line in each graph represent propagation >1-fold.