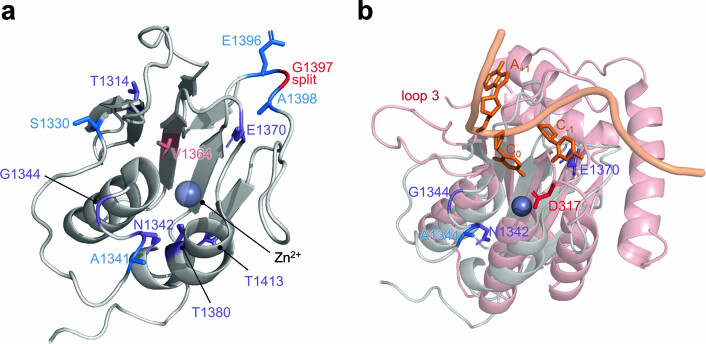
Extended Data Fig. 10. Structural alignment of DddA with ssDNA-bound APOBEC3G.
a, Crystal structure of DddA (grey, PDB 6U08) complexed with DddI immunity protein (not shown). Positions of mutations common to the CCC- and GCC-specific evolutions are colored in purple. Additional mutations are colored according to Fig. 3c. DddA was split at G1397 (red) to generate T7-DdCBE for PANCE and PACE. b, DddA (PDB 6UO8, grey) was aligned to the catalytic domain of APOBEC3G (PDB 2KBO, red) complexed to its ssDNA 5’-CCA substrate (orange) using Pymol. The target C undergoing deamination by APOBEC3G is indicated as C0. Reversion analysis on the DddA11 mutant indicated that A1341V, N1342S and E1370K are critical for expanding the targeting scope of DddA (see Fig. 3f). D317 (red) confers 5’-CC specificity in APOBEC3G and loop 3 controls the catalytic activity of the APOBEC3G.