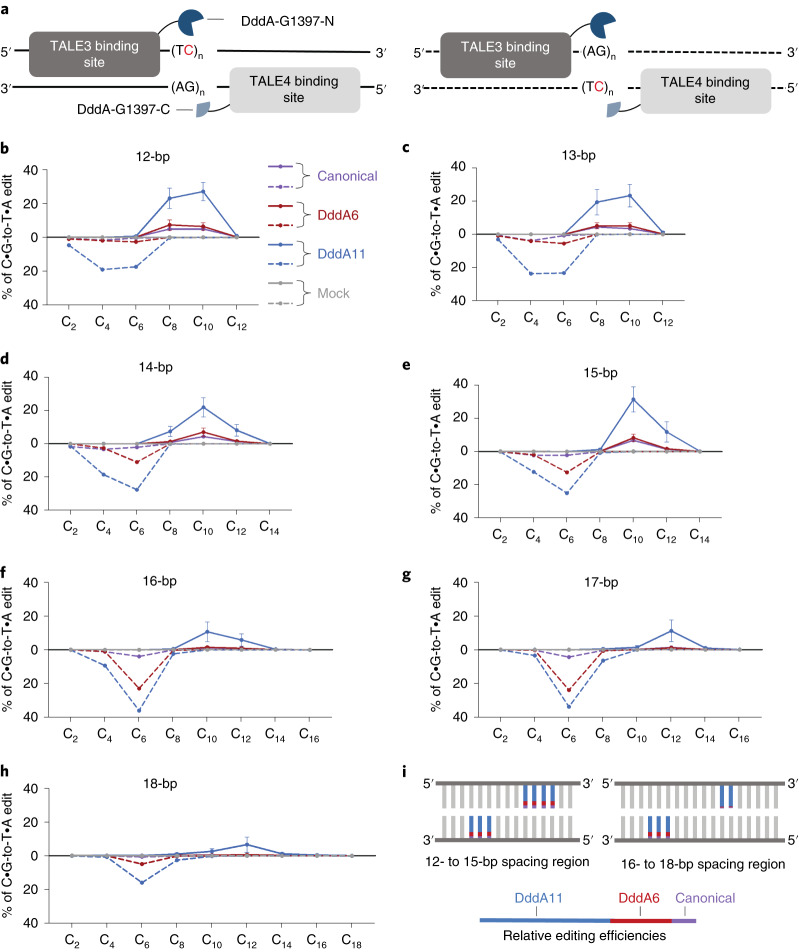
Fig. 4. Editing windows of canonical and evolved T7-DdCBE.
a, Split orientation of T7-DdCBE and its target spacing region. Each spacing region contains TC repeats within the top strand (left, solid line) or bottom strand (right, dashed line). Lengths of spacing regions ranged from 12 bp to 18 bp. b–h, Editing efficiencies mediated by canonical DdCBE (purple), DddA6-containing DdCBE (red) and DddA11-containing DdCBE (blue) are shown for each cytosine positioned within the spacing region length of 12 bp (b), 13 bp (c), 14 bp (d), 15 bp (e), 16 bp (f), 17 bp (g) and 18 bp (h). Subscripted numbers refer to the positions of cytosines in the spacing region, counting the DNA nucleotide immediately after the binding site of TALE3 as position 1. Editing efficiencies associated with the top and bottom strand are shown as solid and dashed lines, respectively. Mock-treated cells contained only the library of target plasmids (gray). For b–h, values and error bars reflect the mean ± s.d. of n = 3 independent biological replicates. i, Approximate editing windows for canonical (purple), DddA6 (red) and DddA11 (blue) variants of T7-DdCBE containing the G1397 split. The length of each colored line reflects the approximate relative editing efficiency for each DddA variant.