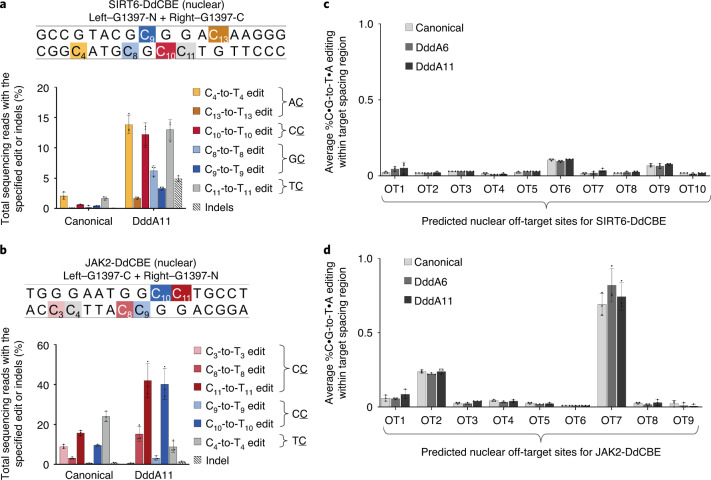
Fig. 5. DddA11 expands targeting scope for nuclear DNA editing.
a, b, Nuclear DNA editing efficiencies of HEK293T cells treated with the canonical or DddA11 variant of SIRT6-DdCBE (a) or JAK2-DdCBE (b). Target spacing regions and split DddA orientations are shown for each base editor. Cytosines highlighted in yellow, red or blue are in AC, CC or GC contexts, respectively. The architecture of each nuclear DdCBE half is bpNLS–2xUGI–4-amino-acid linker–TALE–[DddA half]. bpNLS, bipartite nuclear localization signal. c, d, Average frequencies of all possible C•G-to-T•A conversions within a predicted off-target spacing region associated with SIRT6-DdCBE (a) and JAK2-DdCBE (b). See Supplementary Table 6 for ranking of predicted off-target sites and Supplementary Table 8 for off-target site amplicons. For a–d, values and error bars reflect the mean ± s.d. of n = 3 independent biological replicates.