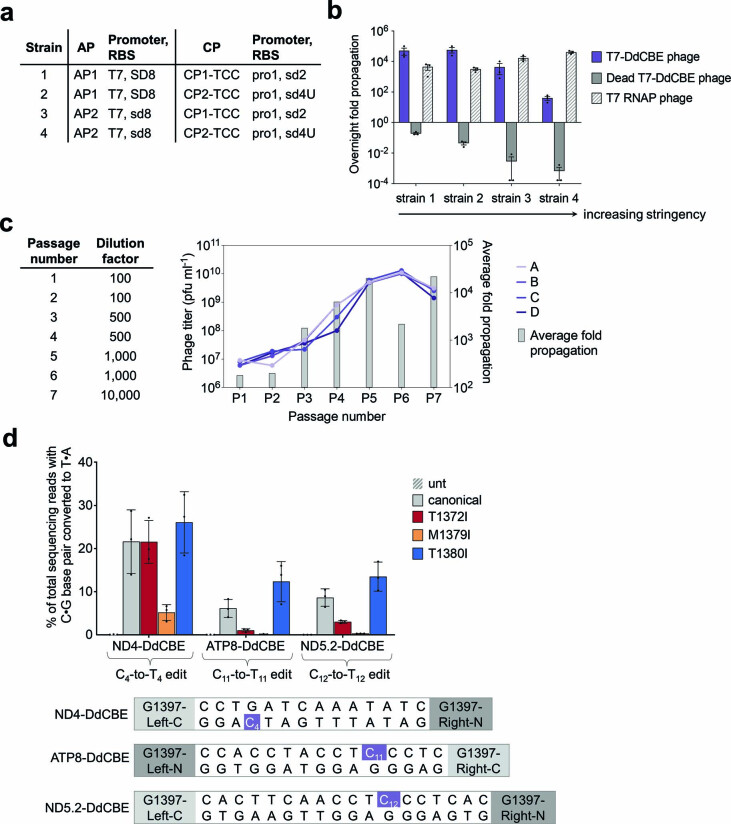
Extended Data Fig. 1. Evolution of canonical T7-DdCBE for improved TC activity using PANCE.
a, Strains for screening selection stringency. Strains were generated by transformation with a variant of an AP and a variant of a CP. All CPs encode a TCC linker. Relative RBS strengths of SD8, sd8, sd2 and sd4U are 1.0, 0.20, 0.010 and 0.00040, respectively. b, Overnight phage propagation of indicated SPs in host strains with increasing stringencies. Dead T7-DdCBE phage contained the catalytically inactivating E1347A mutation in DddA. The fold phage propagation is the output phage titer divided by the input titer. c, Phage passage schedule for canonical T7-DdCBE evolution in PANCE using strain 4 transformed with MP6. Table indicates the dilution factor for the input phage population. Output phage titers for each replicate (A, B, C and D) are shown for each passage. Average fold propagation was obtained by averaging the fold propagation obtained from the four replicates A-D. d, Mitochondrial base editing efficiencies of HEK293T cells treated with canonical DdCBE or with DdCBEs containing the indicated mutations within DddA. For each base editor, the DddA split orientation and target cytosine (purple) within the spacing region is indicated. For b and d, values and error bars reflect the mean±s.d of n = 3 independent biological replicates.