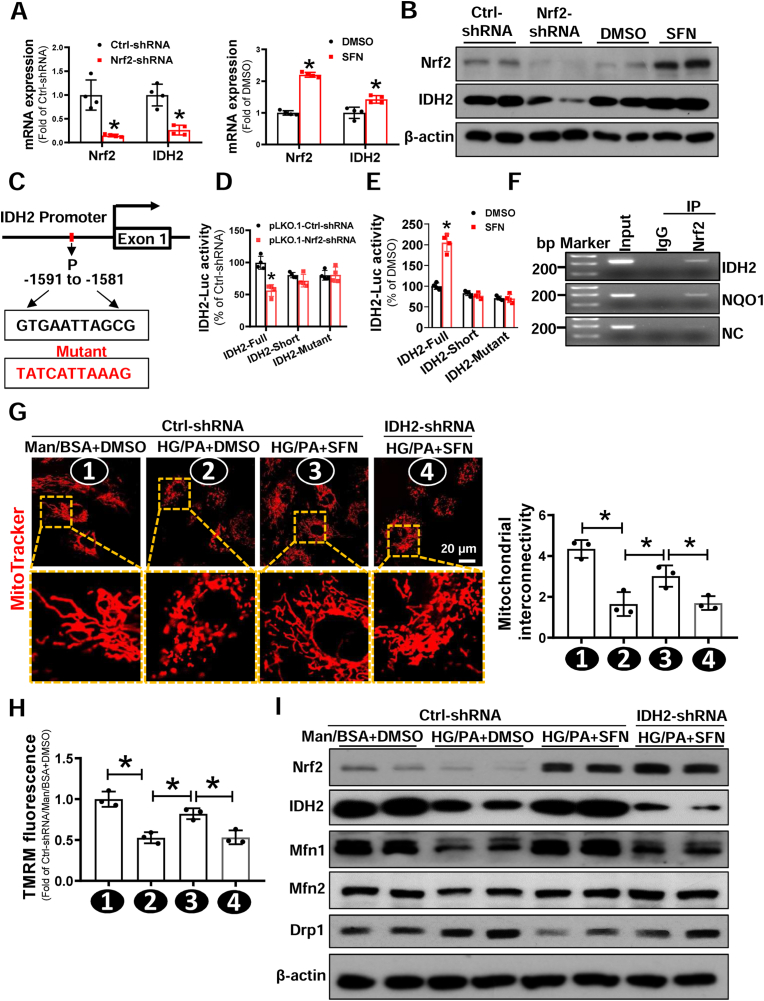
Fig. 7.
Nrf2 suppression of diabetes-induced mitochondrial fission of EPCs is dependent on transcriptional upregulation of IDH2. (A-B) Healthy-EPCs were transfected with lentivirus carrying Ctrl-shRNA or Nrf2-shRNA, or treated with DMSO or Nrf2 activator SFN. The (A) mRNA and (B) protein expression of Nrf2 and IDH2 were determined by Western blot and qRT-PCR, respectively; the quantitative data was normalized by the average of the respective control for each group, and β-actin was used as a loading control. (C) The putative binding sites of Nrf2 on IDH2 promoter was predicted by JASPAR. (D) HEK-293T cells were co-transfected with PLG3.0-IDH2-Luc reporter (Full, Short or mutant) and Nrf2-shRNA or Ctrl-shRNA plasmid. Renilla luciferase reporter plasmid (pRL-TK) was used as an internal control of transfection. Firefly luciferase activity and Renilla luciferase activity in media were measured at 48 h post-transfection. (E) HEK-293T cells were transfected PLG3.0-IDH2-Luc reporter (Full, Short or mutant), then treated with or without SFN, a Nrf2 activator. pRL-TK was used as an internal control of transfection. Firefly luciferase activity and Renilla luciferase activity in media were measured at 48h post-transfection. (F) Chromatin immunoprecipitation (ChIP) analyses using an anti-Nrf2 or a normal rabbit IgG were performed in EPC cells. Primers specific for IDH2, NQO1, or NC (Negative control) were used. (G–I) Healthy-EPCs were transfected with or without lentivirus carrying Ctrl-shRNA or IDH2-shRNA, respectively, followed by exposure to high glucose (HG, 30 mmol/L) plus palmitate (PA, 100 μmol/L) or Ctrl (mannitol plus bovine serum albumin, Man/BSA) in the presence of SFN or DMSO for 24 h. (G) Micrographs of mitochondrial morphology were visualized by MitoTracker Red CMXRos probe staining, and the morphological alterations were quantified by Image J and expressed as mitochondrial interconnectivity. (H) Mitochondrial membrane potential was detected by TMRM staining, and the data was quantified by a flow cytometry and normalized by the average fluorescence density of the Ctrl-shRNA/Man/BSA+DMSO group. (I) The protein expression of mitochondrial fission and fusion related proteins was determined by Western blot, and β-actin was used as a loading control. At least three independent experiments for each study. Data shown in graphs represents the means ± SD. *p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)