Abstract
The yiaKLMNOPQRS (yiaK-S) gene cluster of Escherichia coli is believed to be involved in the utilization of a hitherto unknown carbohydrate which generates the intermediate l-xylulose. Transcription of yiaK-S as a single message from the unique promoter found upstream of yiaK is proven in this study. The 5′ end has been located at 60 bp upstream from the ATG. Expression of the yiaK-S operon is controlled in the wild-type strain by a repressor encoded by yiaJ. No inducer molecule of the yiaK-S operon has been identified among over 80 carbohydrate or derivative compounds tested, the system being expressed only in a mutant strain lacking the YiaJ repressor. The lacZ transcriptional fusions in the genetic background of the mutant strain revealed that yiaK-S is modulated by the integration host factor and by the cyclic AMP (cAMP)-cAMP receptor protein (Crp) activator complex. A twofold increase in the induction was observed during anaerobic growth, which was independent of ArcA or Fnr. Gel mobility shift assays showed that the YiaJ repressor binds to a promoter fragment extending from −50 to +121. These studies also showed that the cAMP-Crp complex can bind to two different sites. The lacZ transcriptional fusions of different fragments of the promoter demonstrated that binding of cAMP-Crp to the Crp site 1, centered at −106, is essential for yiaK-S expression. The 5′ end of the yiaJ gene was determined, and its promoter region was found to overlap with the divergent yiaK-S promoter. Expression of yiaJ is autogenously regulated and reduced by the binding of Crp-cAMP to the Crp site 1 of the yiaK-S promoter.
The gene cluster yiaKLMNOPQRS (yiaK-S) (Fig. 1), labeled according to the latest notation (17) proposed for the genes of unknown function in the Escherichia coli genome (accession no. U00039), lies at min 80.7 of the bacterial chromosome. Sequence similarity studies allowed us to assign a carbohydrate metabolism function to this system (36). Previous work by Sanchez et al. (32) identified the yiaP gene product as a highly specific l-xylulose kinase purified from mutant cells selected for their ability to grow on the rare pentose l-lyxose. However, the yiaK-S operon seems only fortuitously used for the metabolism of the intermediate l-xylulose formed from l-lyxose. The natural origin of l-xylulose may result from the action of the yiaK-S-encoded proteins on the unknown substrate. Of the nine genes in the yiaK-S cluster, recently we have shown that two more gene products are involved in the metabolism of endogenous l-xylulose (14). Gene yiaR is believed to encode a 3-epimerase, while gene yiaS encodes a 4-epimerase. The yiaK-S gene cluster is coordinately regulated by gene yiaJ, located upstream of yiaK-S and divergently transcribed from it. The yiaJ gene product acts as a repressor of yiaK-S. Because no inducer or inducing conditions have been found, expression of this gene cluster has only been detected in mutant strain JA134, selected for its ability to grow on l-lyxose. In this strain, the regulator gene is inactivated by a genome rearrangement mediated by IS1 transposition that leads to constitutive expression of the yiaK-S operon (2). The absence of structural gene transcripts in wild-type strain ECL1 and the absence of regulator transcript in strain JA134 are in accordance with a negative control exercised by the repressor encoded by gene yiaJ. Control of expression of yiaK-S operon by yiaJ and other transcriptional factors is reported here.
FIG. 1.
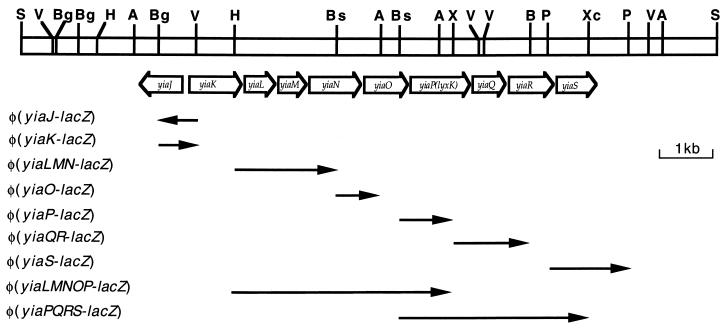
Physical and genetic map of the region encompassing the yiaK-S operon. The bar represents the SalI genome fragment with the relevant restriction sites labeled as follows: A, AgeI; B, BamHI; Bg, BglII; Bs, BstXI; H, HindIII; P, PstI; S, SalI; V, EcoRV; X, XhoI; and Xc, XcmI. Open arrows indicate the extent and direction of transcription of the genes included in the yiaK-S operon. Black arrows represented the fragments fused to lacZ for testing yia promoter function.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
All of the strains used were E. coli K-12 derivatives. The genotype and sources of the relevant bacterial strains are given in Table 1. Genetic crosses were performed by P1vir-mediated transduction (23). Transductants that incorporated the arcA mutation were selected by their sensitivity to O-toluidine blue (15), and those incorporating the fnr mutation were selected by their inability to grow anaerobically on glycerol plus nitrate (37). Transductants that incorporated a himA, himD3, or crp mutation were selected by the linked antibiotic resistance.
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| ECL1 | HfrC phoA8 relA1 tonA22 T2r (λ) | 19 |
| JA134 | ECL1 Lyx+ | 32 |
| CH1828 | araD139 Δ(araBC-leu)7687 ΔlacX74 galU galK hsdR (rK− mK+) rpsL160 thi zce-726::Tn10 rne-50(Ts) | 26 |
| TE2680 | F− λ− IN (rrnD-rrnE) Δ(lac)X74 rplS galK2 recD::Tn10d-tet trpDC700::putPA1303::[Kans Camrlac] | 8 |
| XL1-Blue | recA1 lac endA1 gyrA96 thi hsdR17 supE44 relA1 (F′ proAB lacIqlacZ dM15 Tn10) | Stratagene |
| HN1491 | strA galK2 suo λ− F−himA::cat | 12 |
| K2704 | strA galK2 suo λ− F−himD3::cat | 25 |
| ECL618 | arcA2 zjj::Tn10 | 21 |
| JRG1728 | Δ(tyrR-fnr-rac-trg)17 zdd-230::Tn9 | 37 |
| MC4100 crp | MC4100 crp::cat | 34 |
| JA161 | JA134 yiaJ::cat | 2 |
| JA180 | CH1828 yiaJ::cat | This study |
| JA181 | JA134 yiaK::Tn5 | This study |
| JA182 | JA134 Δ(tyrR-fnr-rac-trg)17 zdd-230::Tn9 | This study |
| JA183 | JA134 arcA2 zjj::Tn10 | This study |
| JA184 | JA134 himA::cat | This study |
| JA185 | JA134 himD3::cat | This study |
| JA186 | JA134 crp::cat | This study |
| JA187 | JA134 cya::Tn5 | This study |
Growth conditions and preparation of cell extracts.
Cells were grown on Luria broth or minimal medium and harvested at the end of the exponential phase as described previously (4). Carbon sources were added to a basal inorganic medium at a 60 mM carbon concentration for aerobic growth and 120 mM for anaerobic growth. For anaerobic respiration, nitrate was also added to the culture at a 20 mM concentration. Casein acid hydrolysate (CAA) was used at 0.5 or 1% depending on oxygen availability. To search for the inducer, a screening of candidate molecules was set up by adding to the CAA medium the following compounds at the carbon concentration indicated above: d-raffinose (C18); d-glucopyranosyl-d-fructose, cellobiose, saccharose, trehalose, and maltose (C12); N-acetylmuramic acid (C11); N-acetylglucosamine (C8); methyl-d-mannopyranoside, pimelic acid, and d-glucoheptonic acid (C7); d-galactose, d-fucose, l-fucose, d-fructose, l-fructose, d-mannose, l-mannose, d-sorbose, l-sorbose, inulin, d-glucose, d-tagatose, d-allose, d-talose, d-psicose, l-rhamnose, d-mannitol, d-sorbitol, myo-inositol, dulcitol, l-fucitol, l-iditol, gluconic acid, galacturonic acid, glucuronic acid, saccharic acid, phthalic acid, l-ascorbic acid, and adipic acid (C6); d-arabinose, l-arabinose, d-xylose, l-xylose, d-lyxose, l-lyxose, d-arabitol, l-arabitol, d-ribose, d-ribulose, d-xylulose, adonitol, xylitol, α-ketoglutaric acid, β-ketoglutaric acid (C5); erythritol, succinic semialdehyde, succinic acid, fumaric acid, d,l-malic acid, tartaric acid, diglycolic acid (C4); d-lactate, l-lactate, d,l-glyceraldehyde, glycerol, pyruvic acid (C3); and glycolic acid and oxalic acid (C2). Citrus fruit pectin and apple pectin were prepared at 0.14% in sodium acetate buffer (pH 4.0) and incubated overnight at 25°C with 400 U of Aspergillus niger pectinase, and the hydrolysis products were diluted 10-fold in the medium. All compounds tested were obtained from Sigma Chemical Co. (St. Louis, Mo.).
When necessary, the antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; tetracycline, 12.5 μg/ml; chloramphenicol, 30 μg/ml; and novobiocin, 200 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) and isopropyl-β-d-thiogalactoside were used at 30 and 10 μg/ml, respectively.
For β-galactosidase assays, the cells were allowed to double five or six times to an A600 of 0.5 for aerobic cultures or 0.25 for anaerobic cultures.
Cell extracts were prepared as described previously (4). Protein concentration was determined by the method of Lowry et al. (20) with bovine serum albumin as a standard. β-Galactosidase activity was assayed by hydrolysis of o-nitrophenyl-β-d-galactopyranoside and expressed as Miller units (23). The data reported are a representative set of at least four separate experiments performed in duplicate.
Northern blot analysis and primer extension.
For preparation of total RNA, cells of a 25-ml culture grown to an A650 of 0.5 were collected by centrifugation at 5,000 × g for 10 min and processed as described by Belasco et al. (3). Northern blot hybridization was performed with each RNA sample (10 μg) by the procedure described by Moralejo et al. (24). For determination of the 5′ end of the structural and the regulatory genes, the following oligonucleotides were used as primers: 5′-GCCGCGTGAAATTAAGACCCG-3′ complementary to an internal region within yiaK and 5′-CTTTTTCCTGTGCCATCTCGTTC-3′ complementary to an internal region within yiaJ. The reactions were performed with 50 μg of total RNA at 37°C for 30 min with 200 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies, Inc.) and [α-35S]thio-dATP (>1,000 Ci/mmol; Amersham Pharmacia Biotech) and followed by a 30-min chase with all four nucleotides (at 1 mM each). As a reference, double-strand sequence reactions were performed with the primers used for the primer extension experiments.
DNA manipulation and genetic techniques.
Plasmid DNA was routinely prepared by the boiling method (13). For large-scale preparation, a crude DNA sample was subjected to purification on a column (Qiagen GmbH, Düsseldorf, Germany). DNA manipulations were performed essentially as described by Sambrook et al. (31). The DNA sequence was determined by using the dideoxy-chain termination procedure of Sanger et al. (33), with double-stranded plasmid as the template.
Tn5 insertion mutagenesis was carried out by infection with phage λ 467 (b221 cIts857 rex::Tn5 Oam29 Pam80), as described by Bruijn and Lupski (5). Tn5 insertion mutants in the yiaK-S gene cluster or in the cya gene were obtained from strain JA134. These mutants were selected for their inability to utilize l-lyxose. The precise location of the insertion in the yiaK gene in strain JA181 and in the cya gene in strain JA187 was assessed by sequencing the chromosomal region close to the Tn5, with an internal sequence of the transposon used as a primer.
Constructions of lacZ fusions and deletions of the yiaK-S promoter.
To create operon fusions, DNA fragments of the 5′-upstream region of some structural genes and the regulator gene were cloned into plasmid pRS550 or pRS551 (35). In some experiments, the DNA fragment was extended to several genes to examine the presence of possible promoters within the coding region (Fig. 1). The pRS plasmids carried a cryptic lac operon and genes that confer resistance to both kanamycin and ampicillin. After introduction of the recombinant plasmids into the tetracycline-resistant strain XL1-Blue, blue colonies on Luria-Bertani plates containing X-Gal, ampicillin, and kanamycin were isolated. Plasmid DNA was sequenced by using the M13 primer to ensure that the desired fragment was inserted in the correct orientation. Single-copy fusions on the E. coli chromosome were obtained by the method of Elliot (8). Plasmids containing the different lacZ fusions were linearized with XhoI or SalI and used to transform strain TE2680. Due to the presence in strain TE2680 of the recD::Tn10 mutation and sequences inserted into the trp operon that are homologous to sequences in pRS plasmids, this strain recombines linear pRS550- or pRS551-based plasmids into its chromosome. The transformants were selected for kanamycin resistance and screened for sensitivity to ampicillin and chloramphenicol. P1vir lysates were made to transduce the fusions into other genetic backgrounds.
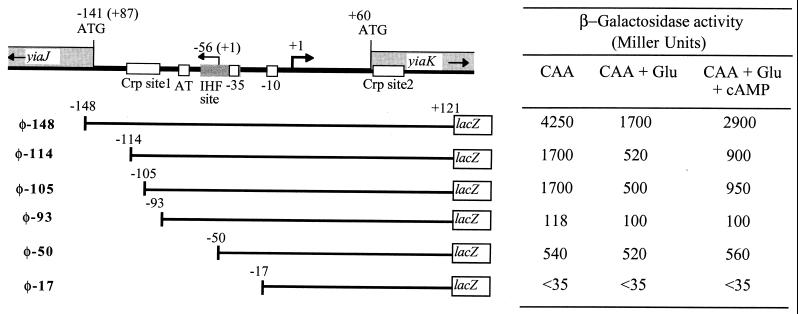
Fragments containing sequences of the promoter region were created by PCR. The 20-mer oligonucleotide 5′-ACGCCGCGTGAAATTAAGAC-3′ identical to the sequence between +121 and +102 of the noncoding strand of gene yiaK was used as the constant primer. This oligomer incorporated eight bases at the 5′ end, which included an EcoRI site at one end of the PCR product. The partner primers extended from the following positions: −148 to −128, −114 to −94, −105 to −85, −93 to −73, −50 to −30, and −17 to +3 (see Fig. 4), all bearing nine additional nucleotides at the 5′ end, which included a BamHI site at the other end of the PCR product. After digestion with BamHI-EcoRI, the PCR products were cloned into pRS550, and the recombinant plasmids were used to construct single-copy lacZ fusions (labeled as φ-148, φ-114, φ-105, φ-93, φ-50, and φ-17) in the background of strain JA134 or ECL1.
FIG. 4.
Deletion analysis of the yiaK-yiaJ promoter region. A set of 5′ deletions of the yiaK promoter was used to generate transcriptional lacZ fusions on the ECL1 and JA134 chromosome. The relevant regulatory elements of the yiaK promoter are shown at the top. Indicated positions correspond to the yiaK coding strand, whereas those corresponding to the complementary yiaJ coding strand are in parentheses. The 5′ ends are marked by an arrow labeled +1. Upstream of the start site of the yiaK-S operon the −10 and −35 consensus, high AT content region, IHF site, and Crp-binding sites are located by the corresponding boxes. Deleted promoter fragments are represented by lines, and the corresponding lacZ fusion is labeled on the left. The 5′ end of each deletion is indicated by the position number to the left. β-Galactosidase activity values are indicated in the table to the right.
DNA binding studies.
For electrophoretic mobility shift assays, three different PCR-amplified fragments were used as probes. These fragments extended, according to the 5′ end determined for the structural genes (see Fig. 3B), from position −148 to position +121, from −50 to +121, and from −148 to −70. After purification from acrylamide gels, fragments were labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; New England Nuclear).
FIG. 3.
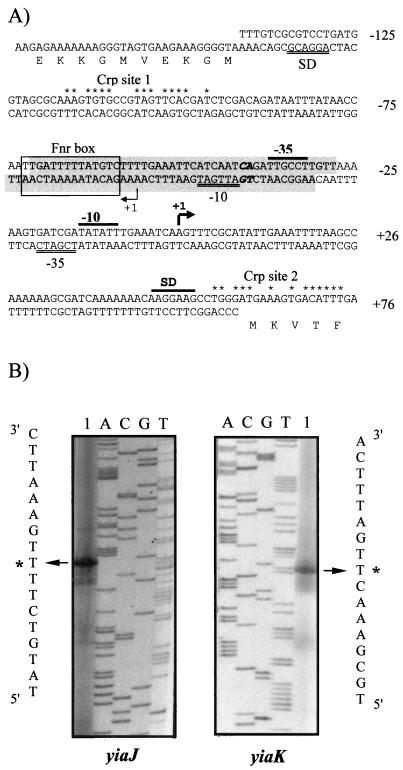
Promoter sequences and primer extension analysis of the divergently transcribed yiaK and yiaJ genes. (A) The sequence containing both overlapping promoters is presented and numbered relative to the 5′ end of the yiaK gene. For each gene, the Shine-Dalgarno sequences (SD) and the −10 and −35 consensus for RNA polymerase binding are indicated by a double underline for yiaJ and a black bar for yiaK. Nucleotides in the extension of the −10 consensus for yiaJ are shown in boldface. The 5′ ends are shown by arrowheads labeled as +1. Potential IHF binding sites, identified by using the MacTargsearch program (10), are shaded. The putative Fnr site is boxed, and positions conserved with respect to the Crp consensus (7) in Crp putative sites are indicated by asterisks. (B) The primed-extended products using total RNA of strain ECL1 (lane 1 of the yiaJ panel) or strain JA134 (lane 1 of the yiaK panel) were electrophoresed with a sequencing ladder (lanes A, C, G, and T) generated by using the same template and primer. A portion of the nucleotide sequence deduced from the sequencing lanes is shown. The most intense extended product assigned as the transcriptional start site for each gene is labeled by an asterisk.
Electrophoretic mobility shift assays were performed with crude extracts obtained as described by Nunoshiba et al. (28). Acrylamide gels containing 10% glycerol were run at 4°C by using 1× Tris-borate-EDTA buffer (1). Protein samples were mixed with 32P-end-labeled DNA substrates (ca. 2.5 nM final concentration, ca. 10,000 to 25,000 cpm) in a 25-μl reaction volume containing 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 10 mM MgCl2, 10% glycerol, and 2 mM dithiothreitol. Poly(dI-dC) was used as a nonspecific competitor. Validation of binding specificity was performed with unlabeled DNA fragments P269, P171, and P78 as competitors for themselves. After incubation for 30 min at 25°C, a 1/6 volume of a 6× gel loading buffer (1) was added, and the mixtures were loaded directly onto prerun gels.
RESULTS
Transcriptional organization of the yiaK-S operon.
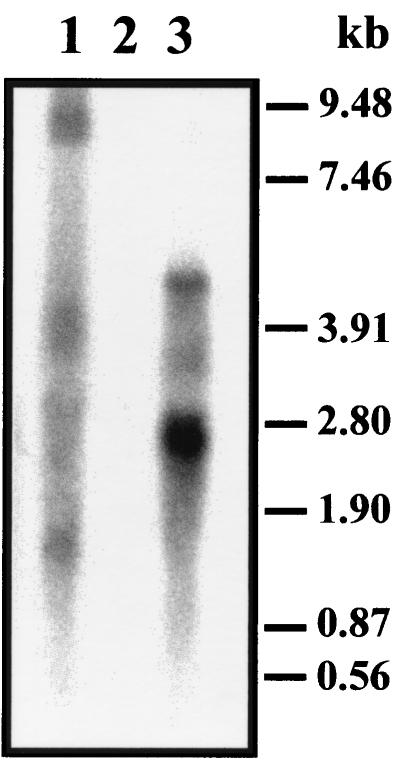
Previous Northern blot experiments with the l-lyxose-positive strain JA134 failed to detect a polycistronic mRNA (2), possibly because of a message decay. To circumvent such a possibility, we transduced the yiaJ::cat of strain JA161 (2) into RNase E temperature-sensitive mutant strain CH1828 (strain JA180). Northern blot hybridization of RNA preparations of mutant strain JA180 grown at a restrictive temperature by using internal probes of yiaP (lyxK), yiaQ, or yiaR showed an mRNA of 8.2 kb corresponding to the full-length transcript of the yiaK-S system. This full-length transcript was not apparent in RNA preparations of strain JA180 grown at a nonrestrictive temperature, while no hybridization bands were observed in a control experiment with RNA of strain CH1828 (Fig. 2).
FIG. 2.
Northern blot of total RNA from strain JA180 grown at 30°C (lane 3), strain JA180 after shifting to 45°C (lane 1), and strain CH1828 grown at 30°C (lane 2). Hybridization was performed with a 0.85-kb yiaR-specific probe.
Transcription of the yiaK-S genes as a single unit was also supported by the properties of lacZ fusions presented in Fig. 1. These operon fusions were transferred to strains ECL1 and JA134. Among these fusions, only the one corresponding to the leading yiaK displayed β-galactosidase activity under any of the growth conditions used, which included CAA in the presence or absence of l-lyxose. Expression of the (yiaK-lacZ) fusion in the genetic background of strain ECL1 resulted in a β-galactosidase activity of 42 Miller units, while its expression in the yiaJ mutant strain JA134 displayed high levels of β-galactosidase activity (4,250 Miller units) in both the absence and the presence of l-lyxose. These observations indicated that the only functional promoter for the structural genes is located upstream of yiaK.
Processing of the yiaK-S genetic cluster as a single transcriptional unit was also consistent with the polarity effects caused by Tn5 insertion mutation in several of the intervening genes, notably in yiaK, the first gene transcribed. Indeed, the yiaK mutant strain JA181 displayed neither l-xylulose kinase activity nor yiaK-S transcript, as shown by the absence of hybridization bands in Northern experiments (data not shown).
As reported previously, the regulator gene yiaJ, divergently transcribed, yielded a transcript of 0.95 kb in RNA preparations of strain ECL1, but not of mutant strain JA134 (2). The operon fusion yiaJ-lacZ corresponding to the putative regulator gene, as is common to other regulator genes, showed a basal level of β-galactosidase activity in strain ECL1, indicating regular function of its promoter (Table 2). The increase in activity level observed in strain JA134, impaired in the regulator gene, could indicate the autogenous regulation of yiaJ expression. Consistently overexpression of YiaJ from pJB1 reduced the level of yiaJ-lacZ expression in strain JA134.
TABLE 2.
β-Galactosidase activities of φ(yiaJ-lacZ) and φ(yiaK-lacZ) in the genetic background of wild-type strain ECL1 and mutant strain JA134 grown in different conditions
| Carbon source | Oxygen | Carried fusion | β-Galactosidase activity in recipient strain (Miller units)
|
|
|---|---|---|---|---|
| ECL1 | JA134 | |||
| CAA | + | φ(yiaJ-lacZ) | 35 | 270 |
| CAA + glucose | + | φ(yiaJ-lacZ) | 175 | 450 |
| CAA + glucose + cAMP | + | φ(yiaJ-lacZ) | 170 | 430 |
| CAA | + | φ(yiaK-lacZ) | 42 | 4,250 |
| CAA + glucose | + | φ(yiaK-lacZ) | <35 | 1,700 |
| CAA + glucose + cAMP | + | φ(yiaK-lacZ) | <35 | 2,900 |
| Succinate | + | φ(yiaK-lacZ) | 46 | 14,500 |
| CAA + ascorbate | + | φ(yiaK-lacZ) | 300 | 4,250 |
| CAA + fucose | + | φ(yiaK-lacZ) | 275 | 4,220 |
| CAA | − | φ(yiaK-lacZ) | 40 | 9,400 |
| CAA + glucose | − | φ(yiaK-lacZ) | <35 | 1,450 |
| CAA + glucose + cAMP | − | φ(yiaK-lacZ) | <35 | 2,940 |
Mapping of the mRNA 5′ end for the structural and regulator transcriptional units.
The 5′ end of the structural genes was determined by primer extension analysis. Total RNA from strain JA134, hence containing structural yiaK-S gene transcripts, was obtained by growth on l-lyxose and also on CAA. For the primer extension reaction, primer complementary to an internal region of yiaK was used, and a single putative start point was determined (Fig. 3B). The mRNA 5′ end was thus located at 60 bp upstream from the ATG codon. The putative start site, position +1 in Fig. 3A, is preceded by a promoter-like sequence that conforms relatively well to the consensus for ς70 RNA polymerase. In this sequence, a −10 TATA box is clearly defined as well as a −35 consensus box found at 17 nucleotides. Analysis of the sequence also showed two putative Crp consensus sites—one centered at −106 (Crp site 1) and the other at +65 (Crp site 2). Using the MacTargsearch program (10), we identified potential integration host factor (IHF)-binding sites extending from −28 to −71 in the coding strand and from −31 to −74 in the complementary strand. A putative Fnr site has been noted centered at −65.5 (Fig. 3A) matching 7 of 10 positions in the Fnr consensus sequence (TTGAT-N4-ATCAA) (39).
Because the regulator gene yiaJ was transcribed opposite to the neighbor structural gene yiaK, the promoter regions of both genes overlapped. Transcription initiation of yiaJ was also determined by the same method with total RNA obtained from strain ECL1, which contains copies of the regulator transcript. In this way, three products were detected by the radioautography. A major one, located at 87 bp upstream of the regulator ATG on the minus strand, was likely to be the 5′ end for the regulator gene (Fig. 3B). The other minor signals were considered to be alternative, secondary, or artifactual. As described by Kumar et al. (18) in other promoters, an extended −10 region, TGATTGAT (Fig. 3A), could compensate for the poor −10 and −35 consensus and the unusual spacing of 21 bp of this weak promoter.
Expression from the functional promoter.
Since we could not identify the inducer, we studied the promoter function in a biochemical background lacking the repressor. Comparison of β-galactosidase activities expressed by the (yiaK-lacZ) fusion in strain ECL1 and strain JA134 allowed us to further characterize the function of this promoter (Table 2). In this way, wild-type cells displayed basal levels of activity when grown on CAA, whereas the mutant strain JA134 gave high constitutive levels of activity when grown on this carbon source, indicating full transcription from its promoter. This is well in agreement with the repressor model of regulation of this system.
Cultures of strain JA134 carrying φ(yiaK-lacZ) grown in the presence of glucose, both aerobically and anaerobically, showed lower β-galactosidase activities, indicating that glucose produced catabolite repression in this promoter. The addition of cyclic AMP (cAMP) to a 5 mM concentration partially recovered the activity in the presence of glucose. The threefold increased expression of φ(yiaK-lacZ) by growth on succinate (Table 2), which is considered a poor carbon source, also confirmed the regulation of this operon by carbon sources. In contrast, the presence of glucose enhanced the promoter function of the regulator, as indicated by the increased β-galactosidase activity of φ(yiaJ-lacZ). No relevant effect of the cAMP addition was observed in this case. This is further supported by the increase in the expression of (yiaJ-lacZ) fusion (510 Miller units) and the reduction of the expression of (yiaK-lacZ) fusion (55 Miller units) in strain JA186 lacking Crp. Thus, yiaK-S transcription is coordinately repressed in the presence of glucose by both the effect of the absence of cAMP-Crp binding to the promoter and the increase in repressor synthesis.
Expression of this operon was twofold higher in cultures of strain JA134 grown in the absence of oxygen, suggesting an aerobic or anaerobic control. To test whether Fnr and/or Arc is involved in the expression of the yiaK-S operon, we transduced an fnr mutation from strain JRG1728 and an arcA mutation from strain ECL618 to JA134 to yield strains JA182 and JA183, respectively. β-Galactosidase analysis showed that neither Fnr nor Arc controls the aerobic or anaerobic expression of this operon (not shown). The influence of DNA supercoiling in this control was studied by adding 0.2 mM novobiocin to CAA cultures at an A600 of 0.5. Assays of β-galactosidase did not show differences after 1 h of incubation with the antibiotic (not shown).
To determine whether IHF influences yiaK-S expression in vivo, we examined the effect of a mutation in himA or in himD3. The himA::cat and the himD3::cat alleles were transferred from strain HN1491 and strain K2704 to a JA134 background by P1 transduction, yielding strains JA184 and JA185. Introduction of each of these IHF mutations produced a fourfold decrease in β-galactosidase activity in these mutant strains compared with that of the isogenic wild type (data not shown). These results suggest that IHF activates yiaK-S expression.
Deletion analysis of the promoter.
To analyze the cis-acting motifs relevant for regulation of the yiaK-S operon and whether the repressor YiaJ is the regulator protein acting in trans, single-copy fusions to the lacZ reporter gene of serially deleted fragments were obtained and introduced in the JA134 background. Twelve constructs were analyzed in cultures grown aerobically in CAA, in both the presence and absence of glucose. Figure 4 displays only those relevant in this analysis. The shortest fusion expressing a level of β-galactosidase activity similar to that of φ(yiaK-lacZ) was φ-148. Constructs φ-114 and φ-105, partially affecting the Crp site 1, expressed β-galactosidase activities 60% lower than that of the full-length promoter construction, whereas in construct φ-93, from which the Crp site 1 had been completely deleted, the activity decreased 98%. In contrast, construct φ-50, which not only lacks the Crp site 1 but also affects the AT-rich sequence and the IHF binding sites, yielded an activity higher than that of φ-93, with a decrease of only 85% with respect to the control. Catabolite repression by glucose and partial reversion by cAMP were apparent for the cells bearing constructs like φ-148, φ-114, or φ-105, which carry all or part of Crp site 1. No effect was seen with constructs φ-93 and φ-50, which lack Crp site 1. Deletion to position −17, affecting RNA polymerase binding, almost abolished promoter function.
Single-copy fusions from φ-148 to φ-50 in the background of strain ECL1, which expresses the repressor protein, showed no β-galactosidase activity, indicating that the regulator binding site is still present in φ-50 (data not shown).
Gel mobility shift assays.
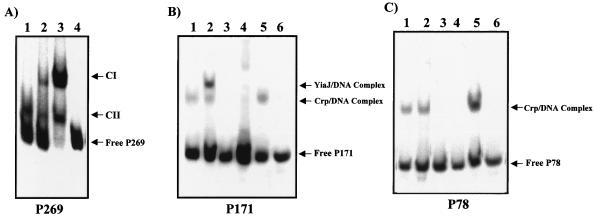
Protein interactions with the yiaK promoter region were assessed by gel mobility shift experiments performed with a 269-bp fragment (positions −148 to +121, labeled P269) belonging to the full-length promoter region. Experiments performed with 10 μg of crude extracts of strain JA134, lacking the repressor, displayed two retarded complexes (CI and CII in lane 3, Fig. 5A). No additional complexes were observed with extracts of strain JA134 transformed with plasmid pJB1 (2), from which YiaJ was overexpressed (lane 2, Fig. 5A). When a crude extract of strain JA184 was used, the CI complex disappeared (lane 1, Fig. 5A), indicating that the formation of this complex was dependent on the IHF binding.
FIG. 5.
Interactions of IHF, Crp, and YiaJ with different fragments of the yiaK promoter region. The experiments were performed with the following 32P-labeled probes: P269 (from −148 to +121), corresponding to the full-length region (A); P171 (from −50 to +121) (B); and P78 (from −148 to −70) (C). The migration patterns of the different DNA-protein complexes are indicated (CI, complex I; CII, complex II). (A) Complexes formed with extracts of strain JA184 (lane 1), strain JA134 previously transformed with plasmid pJB1 (lane 2), strain JA134 (lane 3), and no extract (lane 4). (B and C) Complexes formed with crude extracts of strain JA134 (lanes 1), strain JA134 previously transformed with plasmid pJB1 (lanes 2), strain JA186 (lanes 3), strain JA187 (lanes 4), strain JA184 (lanes 5), and no extract (lanes 6).
To determine the location of the fragment responsible for the repressor binding, the total promoter region was split into two parts: one from −50 to +121 (labeled P171) and the other from −148 to −70 (labeled P78). Neither of the two fragments contained the IHF site, whose occupancy yielded a complex (CI) hiding the other complexes formed by binding of additional proteins. Analyses with extracts of strain JA134 (lane 1 of Fig. 5B and C) or strain JA134 overexpressing YiaJ (lane 2 of Fig. 5B and C) showed, in fragment P171 but not in fragment P78, a retarded complex corresponding to the YiaJ repressor binding.
The identity of the Crp-DNA complexes was approached attending to the previous identification by sequence analysis of two consensus sites for Crp binding in the 269 promoter region. The results presented in Fig. 5B and C show a shifted band with either the P171 or P78 fragment when crude extracts of strain JA134 (lanes 1) or strain JA184 (lanes 5) were used. In contrast, the Crp-DNA bands were absent when crude extracts of Crp mutant strain JA186 (lanes 3) or Cya mutant strain JA187 (lanes 4) were used.
Search for inducers.
As for all genetic systems of unknown function disclosed by the genome sequencing project, identification of the substrate(s) or inducer(s) of the yiaK-S operon is a priority subject. The similarity of these gene sequences and organization to those of other known systems gives some clues for the substrate-candidate structure. In this sense, a similar operon was found in Haemophilus influenzae (6), and the yiaO gene displayed high similarity to a periplasmic solute-binding protein of Rhodobacter capsulatus (9) involved in the transport of dicarboxylates. The YiaJ repressor was also found to have a high similarity to KdgR and Pir of Erwinia chrysanthemi (27, 29), both involved in pectin degradation and classified into the IclR family of regulator proteins. On the other hand, the possibility of assaying candidate inducers with a yiaK-S operon fusion prompted us to approach their identification by using this technique.
Among 80 different compounds tested, only l-fucose and l-ascorbate resulted in modestly increased expression of β-galactosidase activity (Table 2). No induction was seen with any of the other compounds, including pentoses, hexoses, other derivative sugars, mono- and dicarboxylate organic acids, and related compounds listed in Materials and Methods. The possibility of the presence of the inducer in animal gut or lung mucus was explored by using rat specimens suspended in minimal medium and autoclaved. These solutions were added to CAA medium at a final concentration of 0.1%. No induction of β-galactosidase activity was detected by growth in any of these cultures.
The sensitivity of the yiaK-S operon to pH, temperature, osmolarity, oxidative stress, growth phase, and nitrogen availability was also tested. The selected cultures, at an A600 of approximately 0.5, were shocked by addition of NaCl to 0.3 M for osmotic studies (22) or paraquat to 20 mM for oxidative stress studies (30). Heat shock was performed by shifting the culture to 45°C. In these experiments, β-galactosidase activity was assayed after 1 h. For external pH regulation, cells were grown in CAA medium buffered at pH 5.5 or 8.0 as described by Watson et al. (38). For nitrogen limitation, cultures were grown in 1 mM ammonium sulfate. None of these environmental conditions affected the expression of this operon.
As described above, citrus fruit pectin or apple pectin previously hydrolyzed by the action of pectinase did not affect the expression of the structural genes, even in the absence of repressor. However, a 10- to 20-fold induction of φ(yiaJ-lacZ) was observed only in wild-type strain ECL1, but not in mutant strain JA134. At present, we do not know the significance of this effect.
DISCUSSION
Three lines of evidence support the expression of the yiaK-S gene cluster as a single mRNA transcribed from the unique promoter found upstream of yiaK: (i) the presence of full-length transcript in RNase E mutants, (ii) induction of lacZ fusion only from the leading yiaK structural gene, and (iii) polarity effects of yiaK::Tn5 insertion mutation in the yiaK-S transcription. Expression of this operon was previously reported to be under the control of the repressor YiaJ. Consistently, the DNA binding experiments showed that the repressor was bound to a sequence present in promoter fragment P78 (position −50 to +121). This repressor was found to be highly homologous to the IclR family of regulators (32). The cis-acting element recognized by E. coli IclR repressor was shown to be AATTAAAATGGAAATTGTTTTTGATTTTGCATTTT (27), and highly homologous sequences have been reported in E. chrysanthemi for other operator regions recognized by proteins of this family, such as KdgR or Pir (27, 29). In the yiaK-S promoter, the sequence GTTAAAAAGTGATCGATATATTTGAAATCAAGTTT, with many conserved positions (underlined), may also be identified between positions −30 and +5, allowing us to propose it as a putative binding site for the YiaJ repressor.
Our results indicate that the yiaK-S operon is regulated by carbon sources. As shown, succinate increases and glucose reduces its expression in the absence of the repressor. This promoter strongly responds to glucose catabolite repression through the Crp site 1; in contrast, the yiaJ promoter, which overlaps with the yiaK-S promoter and is transcribed divergently from it, responds to the same conditions with increased expression. The mechanism for this yiaJ activation seems to be based on the nonoccupancy of Crp site 1 by the Crp-cAMP complex, hence releasing the obstruction to yiaJ transcription. In this way, catabolite repression of yiaK-S expression is coordinately helped by the increase in the repressor synthesis. In the absence of glucose, Crp-cAMP complex binds to Crp site 1 activating yiaK-S expression and obstructing the yiaJ-encoded repressor expression and synthesis. In spite of the binding observed to Crp site 2, its function remains unknown.
Increased levels of β-galactosidase activity found in anaerobic conditions were not Fnr dependent. The location of the Fnr site at −65.5, different from the −61.5 or −71.5 shown to be the correct distance for activation function (39), may explain the inability of this site to control yiaK-S expression. In this context, examples have been reported in which DNA supercoiling accounts for anaerobic increased expression (16). To study whether this hypothesis applied to our promoter behavior, experiments in the presence of novobiocin were designed. The results indicate that novobiocin did not modify the expression pattern. At present, the regulatory element responsible for this redox control has not been identified.
Sensitivity to IHF was probably due to DNA looping (11) to close the distance between the RNA polymerase site and Crp site 1 or other unidentified cis-acting elements. This effect would be particularly favored by the presence of the AT-rich sequence located between positions −77 and −87.
Up to now, we have been unable to identify the inducer molecule or the conditions required for its induction. One possible explanation would be that the system was a cryptic one activated by a mutation in strain JA134, as proposed in our first report on l-xylulose kinase identification (32). Nevertheless, the normal function of the constitutive promoter and the weak induction by compounds such as l-fucose or l-ascorbate seem to give support to the hypothesis that the yiaK-S operon is normally functional and that the inducing conditions have yet to be found. In this sense, it is of interest to stress that several experiments trying to reproduce the physiological induction conditions have failed. The possibility of mutations in the YiaJ repressor that impaired inducer recognition without affecting DNA binding capacity cannot be ruled out. These mutations would be hardly suppressible, since the recovery of the recognition capacity would be quite unlikely.
Similarities of yiaK-S regulatory systems to KdgR and Pir regulatory elements for the pel regulon in E. chrysanthemi (27, 29) open the possibility of a parallel mechanism as a model for the regulation of these systems. In E. chrysanthemi, the KdgR repressor induces pel genes in response to 2-keto-3-deoxygluconate, and this induction is reinforced by competition with Pir activator. In our case, YiaJ could require the unknown inducing molecule and perhaps the collaboration of another unidentified regulatory protein. Although YiaJ displays more than 70% similarity with Pir, the former acts as a repressor, whereas the latter acts as an activator. The mutation leading to this change in the regulatory function of YiaJ could also be the molecular basis for its inability to recognize the inducer molecule converting the yiaK-S in a cryptic system. Further experiments are in progress to confirm or rule out the crypticity of this genetic system.
ACKNOWLEDGMENTS
This work was supported by grant PB97-0920 from the Dirección General de Enseñanza Superior e Investigación Científica, Madrid, Spain, and partially by the help of the “Comissionat per Universitats i Recerca de la Generalitat de Catalunya.” E.I. is the recipient of a predoctoral fellowship from the Generalitat de Catalunya.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing-Wiley Interscience; 1987. [Google Scholar]
- 2.Badia J, Ibañez E, Sabaté M, Baldomà L, Aguilar J. A rare 920-kilobase chromosomal inversion mediated by IS1 transposition causes constitutive expression of the yiaK-S operon for carbohydrate utilization in Escherichia coli. J Biol Chem. 1998;273:8376–8381. doi: 10.1074/jbc.273.14.8376. [DOI] [PubMed] [Google Scholar]
- 3.Belasco J G, Beatty T, Adams C W, von Gabain A, Cohen S N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985;40:171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- 4.Boronat A, Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol. 1979;140:320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruijn F J, Lupski J R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids. Gene. 1984;27:131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]
- 6.De Rosa R, Labedan B. The evolutionary relationships between the two bacteria Escherichia coli and Haemophilus influenzae and their putative last common ancestor. Mol Biol Evol. 1998;15:17–27. doi: 10.1093/oxfordjournals.molbev.a025843. [DOI] [PubMed] [Google Scholar]
- 7.Ebright R H, Ebright Y W, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the lac DNA site. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliot T. A method for constructing single-copy lac fusions in Salmonella typhimurium and its application to the hemA-prfA operon. J Bacteriol. 1992;174:245–253. doi: 10.1128/jb.174.1.245-253.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golby P, Davies S, Kelly D J, Guest J R, Andrews S C. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J Bacteriol. 1999;181:1238–1248. doi: 10.1128/jb.181.4.1238-1248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrish J A, Schwartz M L, McClure W R. Searching for and predicting the activity of sites for DNA binding proteins: compilations and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 12.Granston B E, Nash H A. Characterization of a set of integration host factor mutants deficient for DNA binding. J Mol Biol. 1993;234:45–49. doi: 10.1006/jmbi.1993.1562. [DOI] [PubMed] [Google Scholar]
- 13.Holmes D S, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 14.Ibáñez E, Giménez R, Pedraza T, Baldomà L, Aguilar J, Badia J. Role of the yiaR and yiaS genes of Escherichia coli in metabolism of endogenously formed l-xylulose. J Bacteriol. 2000;182:4625–4627. doi: 10.1128/jb.182.16.4625-4627.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iuchi S, Furlong D, Lin E C C. Differentiation of arcA, arcB, and cpxA mutant phenotypes of Escherichia coli by sex pilus formation and enzyme regulation. J Bacteriol. 1989;171:2889–2893. doi: 10.1128/jb.171.5.2889-2893.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jamieson D J, Higgins C F. Two genetically distinct pathways for transcriptional regulation of anaerobic gene expression in Salmonella typhimurium. J Bacteriol. 1986;168:389–397. doi: 10.1128/jb.168.1.389-397.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koonin E V, Tatusov R L, Rudd K E. Sequence similarity analysis of Escherichia coli proteins: functional and evolutionary implications. Proc Natl Acad Sci USA. 1995;92:11921–11925. doi: 10.1073/pnas.92.25.11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Grimes B, Fujita N, Makino K, Malloch R A, Hayward R S, Ishihama A. Role of the sigma70 subunit of Escherichia coli RNA polymerase in transcription activation. J Mol Biol. 1994;235:405–413. doi: 10.1006/jmbi.1994.1001. [DOI] [PubMed] [Google Scholar]
- 19.Lin E C C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 20.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Lynch A S, Lin E C C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellies J, Wise A, Villarejo M. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J Bacteriol. 1995;177:144–151. doi: 10.1128/jb.177.1.144-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 24.Moralejo P, Egan S M, Hidalgo E, Aguilar J. Sequencing and characterization of a gene cluster encoding the enzymes for l-rhamnose metabolism in Escherichia coli. J Bacteriol. 1993;175:5585–5594. doi: 10.1128/jb.175.17.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mozola M A, Friedman D I. A phi 80 function inhibitory for growth of lamboid phage in him mutants of Escherichia coli deficient in IHF. Genetic analysis of the Rha phenotype. Virology. 1985;140:313–327. doi: 10.1016/0042-6822(85)90368-x. [DOI] [PubMed] [Google Scholar]
- 26.Mudd E A, Krisch H M, Higgins C F. RNAseE, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 27.Nomura K, Nasser W, Kawaghishi H, Tsuyumu S. The pir gene of Erwinia chrysanthemi EC16 regulates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc Natl Acad Sci USA. 1998;95:14034–14039. doi: 10.1073/pnas.95.24.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunoshiba T, Hidalgo E, Amábile-Cuevas C F, Demple B. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol. 1992;174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reverchon S, Nasser W, Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 30.Rocha E D, Smith C J. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J Bacteriol. 1997;179:7033–7039. doi: 10.1128/jb.179.22.7033-7039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sanchez J C, Gimenez R, Schneider A, Fessner W-D, Baldomà L, Aguilar J, Badia J. Activation of a cryptic gene encoding a kinase for l-xylulose opens a new pathway for the utilization of L-lyxose by Escherichia coli. J Biol Chem. 1994;269:29665–29669. [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawers G, Kaiser M, Sirko A, Freundich M. Transcriptional activation by FNR and CRP: reciprocity of binding-site recognition. Mol Microbiol. 1997;23:835–845. doi: 10.1046/j.1365-2958.1997.2811637.x. [DOI] [PubMed] [Google Scholar]
- 35.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 36.Sofia H J, Burland V, Daniels D L, Plunkett G, Blattner F R. Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiro S, Guest J R. Regulation and over expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987;133:3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- 38.Watson N, Dunyak D S, Rosey E L, Slonczewski J L, Olson E R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992;174:530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wing H J, Williams S M, Busby S J W. Spacing requirements for transcription activation by Escherichia coli FNR protein. J Bacteriol. 1995;177:6704–6710. doi: 10.1128/jb.177.23.6704-6710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]