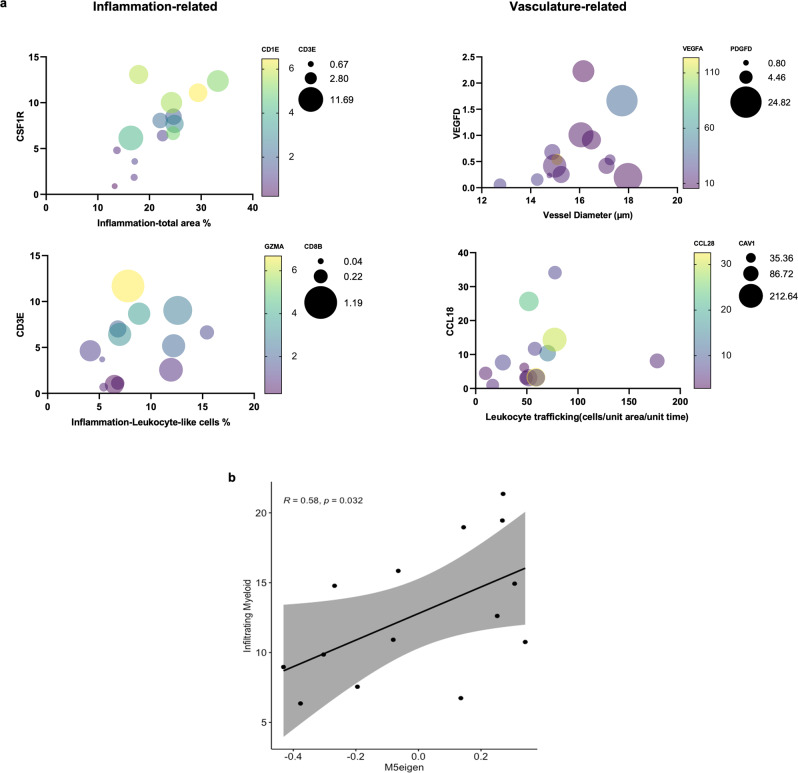
Fig. 6. Quantified RCM TiME features correlate with gene expression.
a Automated features correlated with corresponding gene expression in n = 14 BCC lesions shows moderate to high correlation between total inflammation area with CSF1R (r = 0.73, CI: 0.32 to 0.91, p-value = 0.002), CD1E (r = 0.64, CI: 0.15 to 0.87, p-value= 0.008) and CD3E (r = 0.51, CI:−0.04 to 0.82, p-value = 0.032) and between total leukocyte-like cells area with CD3E (r = 0.6, CI: −0.13 to 0.79, p-value = 0.013), CD8B (r = 0.6, CI: 0.1 to 0.86, p-value = 0.012) and GZMA (r = 0.53, CI: −0.01 to 0.83, p-value = 0.026). Similarly, vessel diameter and leukocyte trafficking were correlated with VEGFD (r = 0.459, CI: −0.1 to 0.80, p-value = 0.050), VEGFA (r = −0.477, CI: −0.81 to 0.09, p-value = 0.044), PDGFD (r = 0.538, CI: 0 to 0.84, p-value= 0.025), and trafficking with CCL18 (r = 0.561, CI: 0.019 to 0.84, p-value= 0.042), CAV-1 (r = 0.468, CI: −0.10 to 0.80, p-value= 0.046) and CCL28 (r = −0.42, CI: −.016 to 0.78, p-value= 0.137), respectively. Non-parametric two-tailed Spearman correlation was computed across each dataset. b Automated features correlated with gene co-expression modules show total myeloid cells on RCM (dendritic cells+macrophages) were significantly correlated with eigengene values for M5 module(Spearman method, p-values estimated using t-distributions). Confidence interval at 95% indicated in gray along with linear regression line and correlation coefficient (p-value = 0.032). Source data are provided as a Source Data file. CSF1R: colony stimulating factor 1-receptor; CD: cluster of differentiation; GZMA: granzyme A; VEGF: vascular endothelial growth factor; PDGFD: platelet derived growth factor D; CCL: CC-chemokine ligand; CAV: caveolin.