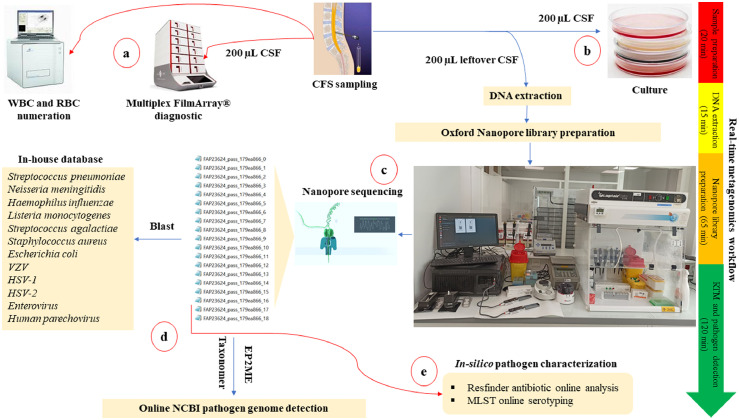
Figure 1.
CSF workflow for the diagnosis of community-acquired meningitis in the POC laboratory for the 52 CSF series prospective investigation. When the CSF sample was received at the POC laboratory, several tests were performed to detect the meningitis causative agent. a) Systematically, the emergency multiplex BioFire FilmArray® assay performed using 200 µL CSF when the sample was received, followed by quantification of the blood cells. b) As per routine diagnosis, all CSF samples received at the POC were routinely cultured. c) The RTM diagnosis was performed in a total workflow that did not exceed four hours. Sample preparation and DNA extraction from left over CSF samples took 35 min, Oxford Nanopore library preparation took 65 min, and the MinION library sequencing took two hours. d) Real-time genome identification was performed directly by blast of the MinION generated data against an in-house database, then against the NCBI database using EPI2ME and Taxonomer online software. e) Antibiotic resistance and pathogen genotyping were in-silico predicted using the ResFinder online database (https://cge.cbs.dtu.dk/services/ResFinder-4.1/), and MLST online database (https://pubmlst.org/organisms).
Abbreviations: CSF: cerebrospinal fluid. WBC: white blood cell. RBC: red blood cell. RTM: real-time metagenomics. MLST: Multi-Locus Sequence Typing.