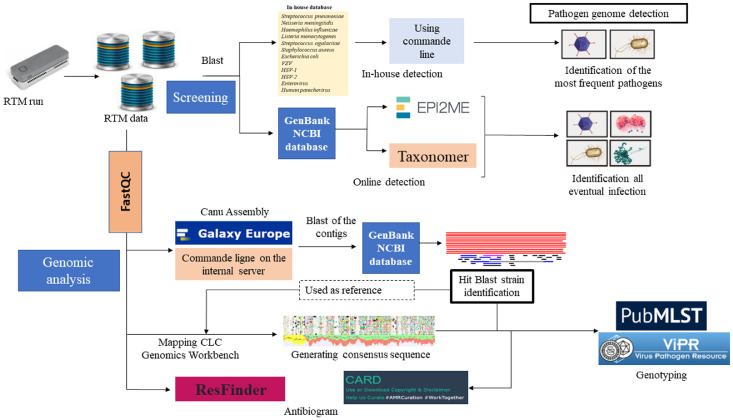
Figure 2.
Bioinformatic pipeline. Rapid pathogen genome identification performed by direct alignment of MinION reads with an in-house database using blastn command line on the IHU server, further against NCBI GenBank by Taxonomer and EPI2ME online software. The analysis was interpreted as positive when ≥ 2 pathogen-specific reads were identified. To confirm pathogen identification, the quality of MinION reads was controlled by FastQC before assembly by “Canu assembler” on Galaxy Europe online software (https://usegalaxy.eu/). Hit-blast strains were identified by Blastn of the generated contigs against GenBank database, then used as reference genome for mapping the total MinION reads by CLC Genomics Workbench software version 21.0.3 (Qiagen). The consensus genomes were extracted in fasta files for pathogens genotyping on MLST on PubMLST (https://pubmlst.org/organisms) for bacteria genotyping and on ViPR online database (https://www.viprbrc.org/) for virus genotyping. The in-silico antibiogram was predicted on ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/) and Resistance Gene Identification (https://card.mcmaster.ca/analyze/rgi) platforms using the total MinION reads and the generated fasta sequences with default settings.
*In-silico antibiogram and genotype were derived from genome sequence only in the case of > 1% genome coverage.