Figure 1. An inventory of parental-specific expression from single mouse embryos.
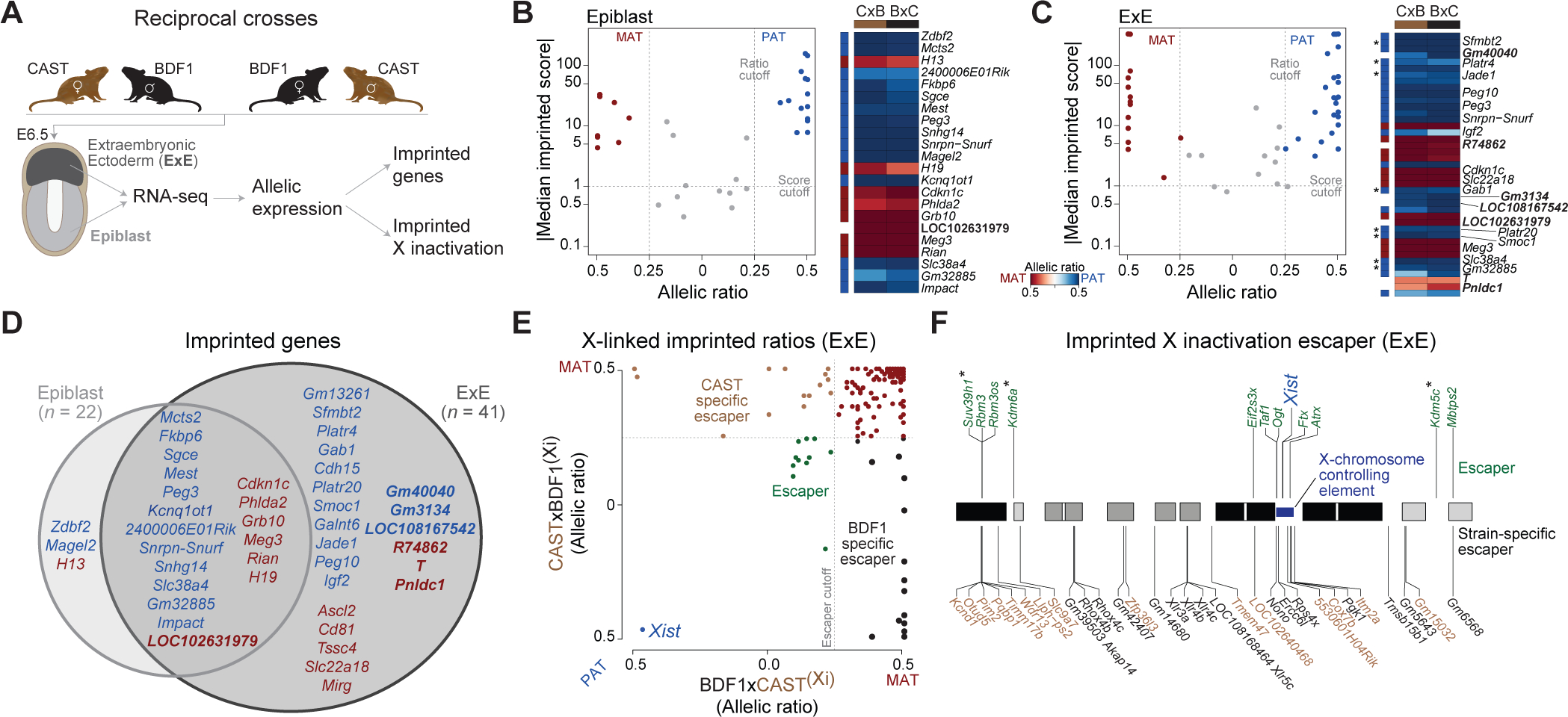
(A) Simplified schematic of our experimental system to obtain parent-specific expression landscapes (imprinting and X inactivation). E6.5 epiblast (light grey) and extraembryonic ectoderm (ExE, dark grey) are isolated from F1 reciprocal crosses (n = 11; 7 BDF1xCAST, 4m/3f and 4 CASTxBDF1, 1m/3f) and subjected to RNA-seq. Colors are used in figures throughout the manuscript to highlight the tissue of origin. (B-C) Imprinted genes identified in the E6.5 Epiblast and ExE (red: maternally expressed, blue: paternally expressed) using a median imprinted score cutoff and allelic ratios of 1 and 0.25, respectively (dashed lines). Corresponding heatmaps show the allelic ratios in the forward (BDF1xCAST) and reverse (CASTxBDF1) crosses. Allelic ratios are adjusted from an initial range of 0 to 1 such that 0 corresponds to equivalent expression between both alleles: 0 = Biallelic, 0.5 = 100% expressed from one allele (MAT (maternal), PAT (paternal), BDF1, or CAST). Previously uncharacterized imprinted genes are indicated in white, including maternal-specific expression of Brachyury (T) within the ExE. A more detailed explanation of the allelic ratio calculation is provided in the Methods. Asterisk indicates known non-canonical imprinting genes (see Table S1 sheet E)
(D) Overlap of imprinted genes between E6.5 Epiblast and ExE (red: maternally expressed, blue: paternal expressed).
(E) Scatter plot showing the allelic ratios for X-linked genes in ExE between forward (BDF1xCAST) and reverse (CASTxBDF1) crosses. Maternally expressed genes (red), XCI escaper genes (green), and strain-specific escape from the CAST (brown) and BDF1 (black) inactive X chromosome are indicated. The dashed line indicates the 0.25 allelic ratio threshold used to determine escaper genes.
(F) Chromosomal overview of genes that maintain biallelic expression on the Xi (“escapers”) including their shared or strain-specific status. Asterisk indicates escaper genes that function as chromatin modifiers.
