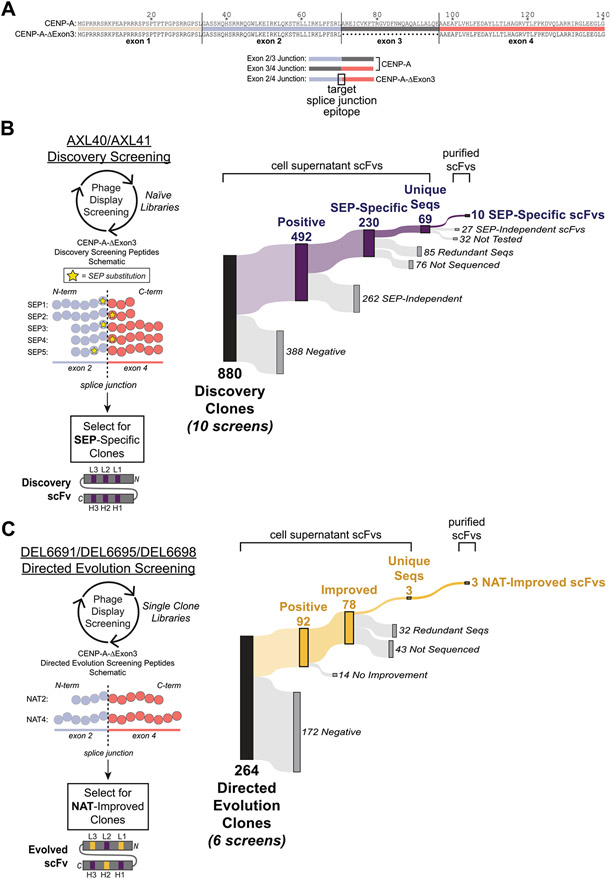
Fig. 1. Peptide designs and screening results for in vitro derivation of splice junction-specific Abs using a nnAA targeting strategy and directed evolution.
(A, top) Sequence alignment of the canonical human CENP-A and the alternatively spliced human CENP-A-ΔExon3 that lacks exon 3. (A, bottom) Schematics of exon-exon junctions that differ between CENP-A and CENP-A-ΔExon3. The exon 2/4 splice junction specific to CENP-A-ΔExon3 is the target epitope for Ab derivation. (B, left) Five Discovery screening peptides (SEP1–5) spanning the exon 2/4 splice junction of CENP-A-ΔExon3 were synthesized with a single non-native SEP substitution (yellow star) at variable positions across the splice junction. Amino acids are denoted by a circle color coded as per (A). Note that total amino acid length is not shown but the relative position of SEP is depicted. Using two different naïve phage display libraries (AXL40 and AXL41), Discovery screening was performed against the five SEP peptides to select for SEP-specific scFvs. A Discovery scFv schematic is shown with CDRs in purple denoting a SEP-specific paratope. (B, right) Summary of Discovery screening results including all 10 unique screens (i.e., each Discovery library against each SEP peptide). “Positive” hits bind to the SEP peptide in ELISA and are further differentiated based on their ability (“SEP-Independent”) or inability (“SEP-Specific”) to bind to the nonphosphorylated, native sequence peptide. Results for each independent screen are presented in Supplementary Table S2. (C, left) Three SEP-specific scFvs were mutagenized and individual Directed Evolution libraries were generated (DEL6691, DEL6695 and DEL6698). Using each library, Directed Evolution screening was performed against the nonphosphorylated sequence peptides [NAT2 and NAT4, color coded as per (A-B)] to select for scFvs with improved binding strength to the native exon 2/4 splice junction of CENP-A-ΔExon3. Note that NAT2 is identical in sequence and length to SEP3–5 (B, left) except for the SEP residue. Each library was screened under two conditions as per Materials and Methods. An example Directed Evolution scFv schematic is shown with preserved parental CDRs in purple and mutated CDRs in yellow denoting an evolved paratope to the native sequence epitope. Note that mutations can be acquired in any of the six CDRs. (C, right) Summary of Directed Evolution screening results including all six unique screens. “Positive” hits bind to the NAT4 peptide in ELISA and are further differentiated based on their ability (“Improved”) or inability (“No Improvement”) to bind to NAT4 stronger than their SEP-specific parental Discovery clone. Results for each independent screen are presented in Supplementary Table S3B.