Fig. 3. Directed Evolution clones demonstrate specificity for the exon 2/4 splice junction of CENP-A-ΔExon3 in split-splice junction ELISAs.
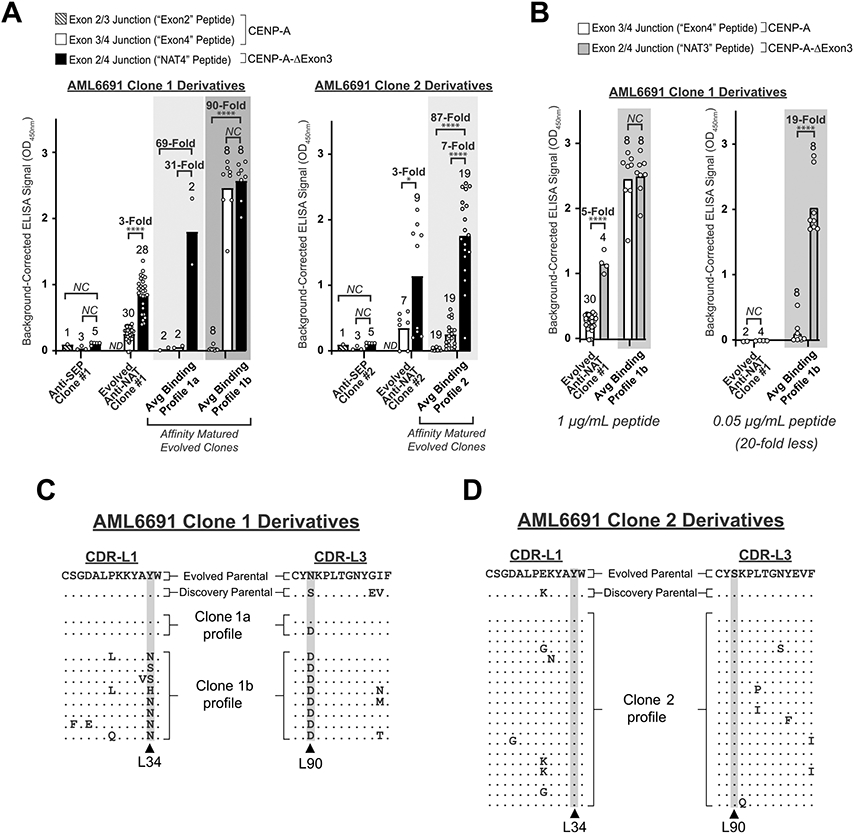
(A) Cell supernatant ELISA graphs comparing splice junction specificity between the Discovery anti-SEP parental scFv (see also Fig. 2A, left), the Directed Evolution anti-NAT parental scFvs (see also Fig. 2A, right), and the affinity matured Directed Evolution anti-NAT scFvs for AML6691 Clone 1 (left) and AML6691 Clone 2 (right). Three CENP-A-ΔExon3 splice junction-spanning peptides were tested: “NAT4” = exon 2/4 splice junction, “Exon2” = exon 2/3 splice junction, and “Exon4” = exon 3/4 splice junction as per Fig. 1A. Each white circle within each bar graph for the Anti-SEP and Evolved Anti-NAT clones represents a biological replicate for scFv expression and ELISA result; bar height represents the average ELISA result of biological replicates. Each white circle within each bar graph for the three “Avg Binding Profile” graphs represents a unique scFv, some of which have biological replicates (Supplementary Figs. S1 and S2) and the average is reported herein; each bar height represents the average ELISA result for the unique scFvs within that group. ND = No Data. NC = No Change. P-values (* = < 0.05, ** = < 0.01, *** = < 0.001, **** = < 0.0001) were calculated using an unpaired t test with Welch’s correction on square root OD450 values in GraphPad Prism on samples with at least two replicates. (B) Cell supernatant ELISA graphs for the AML6691 Clone 1 derivatives against 1 μg/mL (~400 nM) (left) and 0.05 μg/mL (~20 nM) (right) NAT3 (exon 2/4 splice junction) and Exon4 peptides. White circles, bar heights and p-values are defined as in (A). (C and D) CDR-L1 and CDR-L3 protein sequence alignments between the Directed Evolution parental scFv (line 1), the anti-SEP Discovery parental scFv (line 2), and the affinity matured progeny. An enrichment of mutations was identified in the AML6691 Clone 1 affinity matured progeny at Kabat positions L34 and L90 (C), but not in the AML6691 Clone 2 affinity matured progeny (D). No significant enrichment of mutations was identified in any region of the heavy or light chains of the AML6691 Clone 2 progeny.
