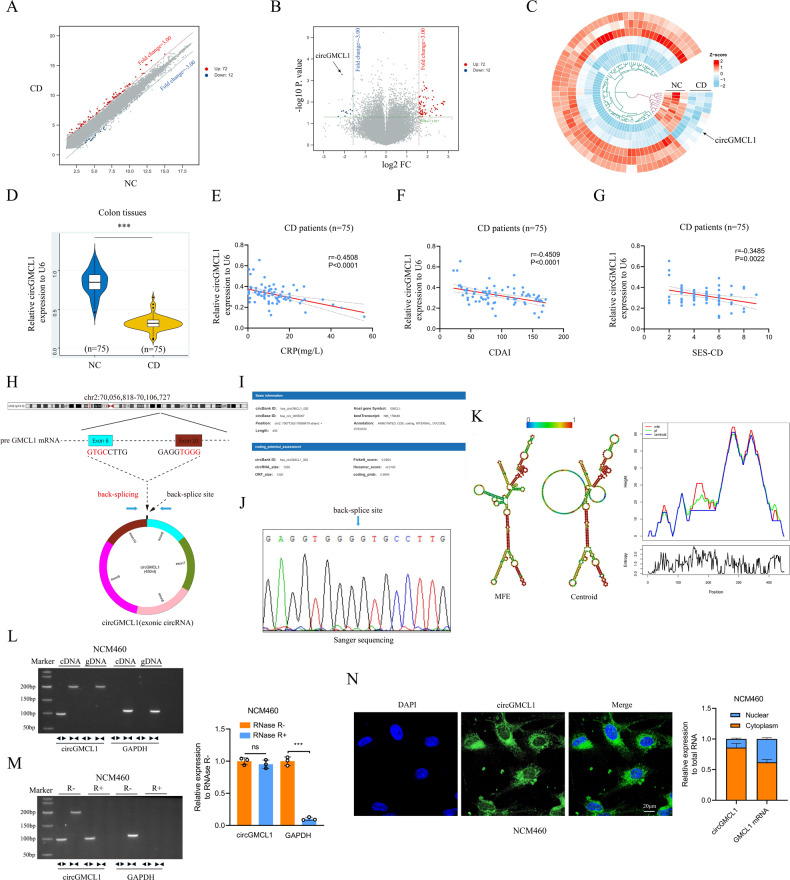
Fig. 1. Identification of circGMCL1 as the candidate circRNA.
Scatter plots A volcano plots B and heatmap C showing circRNA expression profile in colon tissues from CD patients and NCs (threshold: FC > 3, P < 0.05). D Relative circGMCL1 expression in colon tissues from CD patients and NCs as determined by qRT-PCR analysis. Relationship between circGMCL1 expression and CD-associated CRP E, CDAI F and SES-CD G in CD patients (n = 75). H The circle structure of circGMCL1 back-spliced by exon 6 and exon 10. I The basic information of circGMCL1. J The splicing of circGMCL1 by Sanger sequencing in NCM460 cell lines. K The predicted secondary structures of circGMCL1. L Nucleic acid electrophoresis for circGMCL1 in NCM460 cell lines by RT-PCR. M RNase R experiments by nucleic acid electrophoresis and qRT-PCR analyses. N FISH and qRT-PCR analyses performed to reveal the localization and expression of circGMCL1 in NCM460 cells. circRNA, circular RNA; CD, Crohn’s disease; NC, normal control; FC, fold change; qRT-PCR, quantitative real time-polymerase chain reaction; CRP, C-reactive protein; CDAI, Crohn’s disease activity index; SES-CD, simple endoscopic score for Crohn’s disease, MFE minimum free energy, FISH fluorescence in situ hybridization. Data are presented as mean ± SD. ***P < 0.001 by Student’s t tests.