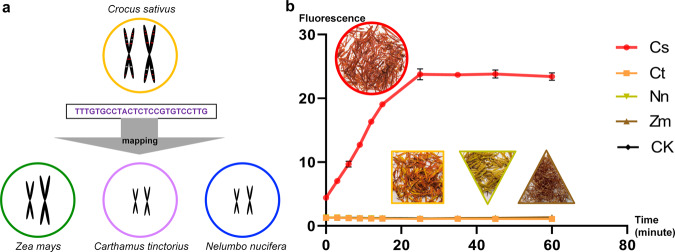
Fig. 4. Identification of Crocus sativus with GAGE.
a Selection of target sequences. The red lines represented Cs_target1 and its copies and white lines represented the sequences within three base mismatches compared to Cs_target1. b Results of the identification of Cr. sativus with GAGE. The plant materials in the circle, square, triangle, and inverse triangle are the stigma of saffron, the flower of safflower, the stamen of lotus, and the style and stigma of corn, respectively. The DNA substrates of each group were as follows: Cs: ITS2 fragments of Cr. sativus; Ct: ITS2 fragments of Ca. tinctorius; Nn: ITS2 fragments of N. nucifera; Zm: ITS2 fragments of Z. mays and CK (negative control): nuclease-free water. All plots data represent means +/− standard deviation (SD) from three independent replicates.