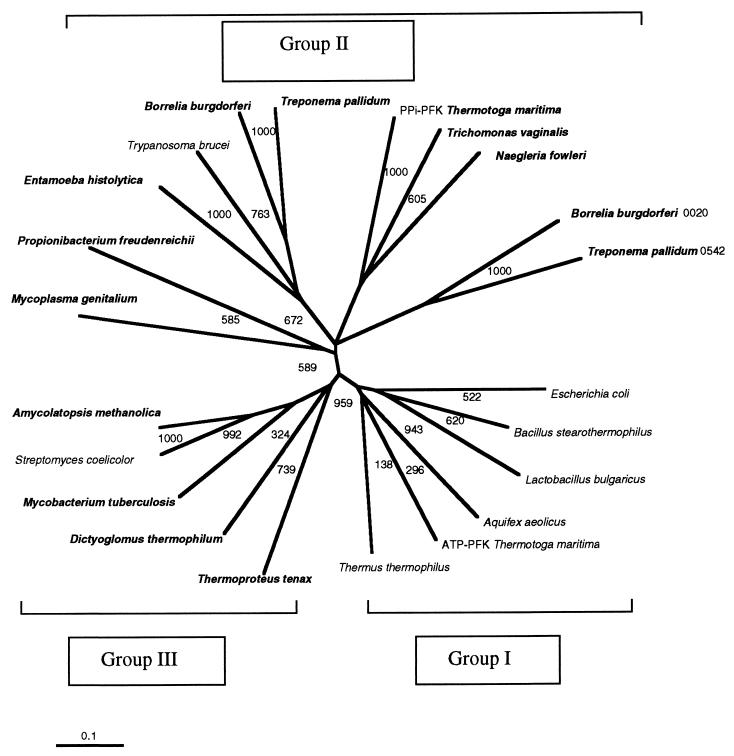
FIG. 2.
Phylogenetic relationships of PPi- and ATP-PFKs. The tree is based on distance analysis (neighbor-joining method) of full amino acid sequences of PPi- and ATP-PFKs in the EMBL and Swiss Prot databanks. Bootstrap values are based on 1,000 replicates and are given at each node. All of the PPi-PFKs are bolded. Group I includes Thermus thermophilus, Thermotoga maritima (ATP-PFK), Aquifex aeolicus, Lactobacillus bulgaricus, B. stearothermophilus, and E. coli. Group II includes Treponema pallidum 0542, Borrelia burgdorferi 0020, Naegleria fowleri, Trichomonas vaginalis, Thermotoga maritima (PPi-PFK), Treponema pallidum, B. burgdorferi, Trypanosoma brucei, Entamoeba histolytica, Propionibacterium freudenreichii, and Mycoplasma genitalium. Group III includes Thermoproteus tenax, D. thermophilum, Mycobacterium tuberculosis, A. methanolica, and S. coelicolor.