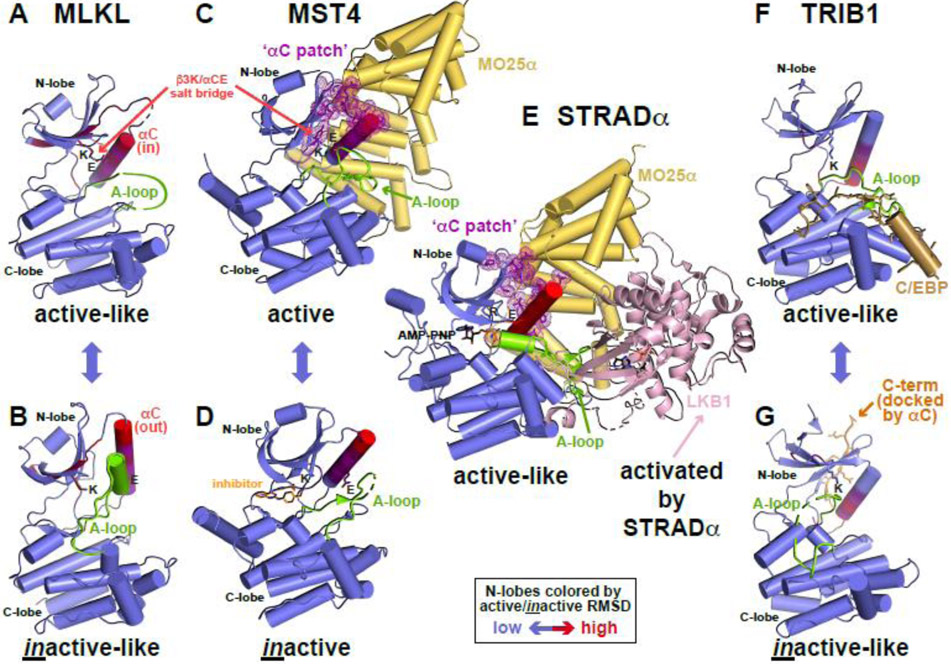
Figure 2. Conformational toggling of pseudokinases.
(A,B) Conformational transition between active-like (A) and inactive-like (B) conformations of MLKL (PDB IDs: 7JW7 [47] and 7JXU [47]), labeled and colored as in Figure 1, with N-lobe colored according to the RMSD between states in an N-lobe overlay.
(C) Allosteric activation of the bona fide protein kinase MST4 by MO25α (PDB ID: 4FZA [48]), resembling EGFR and CDK2 activation in Figure 1. MO25α (yellow) binds to residues in the MST4 N-lobe that constitute the αC patch (C), which in turn coincides with the most variable region in the MST4 N-lobe between active and inactive conformations.
(D) MST4 kinase domain in an inactive conformation (PDB ID: 3GGF [106]), bound to a quinazoline inhibitor [106], which is shown in orange.
(E) Conformational modulation of the pseudokinase STRADα by MO25α to activate LKB1. MO25α binds the αC patch of STRADα to stabilize it in the active-like ATP-bound conformation. The two molecules form a heterotrimer with the bona fide kinase LKB1/STK11 (pink) with STRADα recognizing LKB1 like a pseudosubstrate and allosterically activating it. PDB ID: 2WTK [35].
(F,G) Conformational transition between active-like (F) and inactive-like (G) TRIB1 (PDB IDs: 5CEM [52] and 6DC0 [51]), as shown for MLKL in (A). In the active-like state (F), TRIB1 binds the degron of C/EBP family transcription factors (shown in sand color) like a pseudosubstrate, and promotes release of the C-terminal fragment of TRIB1 (orange in G) that docks adjacent to αC – near the αC patch – in the inactive-like conformation.